| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,469,138 – 11,469,226 |
| Length | 88 |
| Max. P | 0.622222 |

| Location | 11,469,138 – 11,469,226 |
|---|---|
| Length | 88 |
| Sequences | 11 |
| Columns | 89 |
| Reading direction | forward |
| Mean pairwise identity | 81.68 |
| Shannon entropy | 0.40053 |
| G+C content | 0.33220 |
| Mean single sequence MFE | -16.43 |
| Consensus MFE | -10.10 |
| Energy contribution | -10.07 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.622222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11469138 88 + 23011544 -UGACGCUGUUGUGACCUGUCAUUAAACAAAUGACGGCUCAUUUCCAAAUUUGAACCGAGUAAUUUUGCUUAAUUUGUAAUAUUUUUUG -.......(..((((.((((((((.....)))))))).))))..)((((((......(((((....)))))))))))............ ( -15.40, z-score = -1.27, R) >droPer1.super_1 8374893 86 + 10282868 -UGGCAUCGCUGUGACCUGUCAUUAAACAAAUGACGGCUCAUUCCCAAAUUUGUUCCGAGUAAUUUUGCUUAAUUUGUAAUAUUUGC-- -..(((..(..((((.((((((((.....)))))))).))))..)((((((......(((((....))))))))))).......)))-- ( -16.20, z-score = -0.76, R) >dp4.chr4_group3 6924551 86 + 11692001 -UGGCAUCGCUGUGACCUGUCAUUAAACAAAUGACGGCUCAUUCCCAAAUUUGUUCCGAGUAAUUUUGCUUAAUUUGUAAUAUUUGC-- -..(((..(..((((.((((((((.....)))))))).))))..)((((((......(((((....))))))))))).......)))-- ( -16.20, z-score = -0.76, R) >droAna3.scaffold_12916 12173074 82 - 16180835 -------GACUGUGACCUGUCAUUAAACAAAUGACGGCUCAUUUCCAAAUUCGUAGCGAGCAAUUUUGCUUAAUUUGUAAUAUUUUUUG -------((..((((.((((((((.....)))))))).)))).))((((((...((((((....)))))).))))))............ ( -19.10, z-score = -2.91, R) >droEre2.scaffold_4929 12671443 85 - 26641161 ----CGUUGUUGUGACCUGUCAUUAAACAAAUGACGGCUCAUUUCCAAAUUUGAACCGAGUAAUUGUGCUUAAUUUGUAAUAUUUUUUG ----....(..((((.((((((((.....)))))))).))))..)((((((.....(((....))).....))))))............ ( -16.40, z-score = -1.61, R) >droYak2.chr2L 7870487 85 + 22324452 ----CGCUGUUGUGACCUGUCAUUAAACAAAUGACGGCUCAUUUCCAAAUUUGAACCGAGUAAUUUUGCUUAAUUUGUAAUAUUUGUUG ----....(..((((.((((((((.....)))))))).))))..)(((((.((((..(((((....)))))..))))....)))))... ( -16.00, z-score = -1.53, R) >droSec1.super_3 6863896 82 + 7220098 -------UGUUGUGACCUGUCAUUAAACAAAUGACGGCUCAUUUUCAAAUUUGAACAGAGUAAUUUUGCUUAAUUUGUAAUAUUUUUUG -------((..((((.((((((((.....)))))))).))))...))........(((((.(((.((((.......)))).)))))))) ( -16.30, z-score = -1.83, R) >droSim1.chr2L 11280551 82 + 22036055 -------UGUUGUGACCUGUCAUUAAACAAAUGACGGCUCAUUUCCAAAUUUGAACCGAGUAAUUUUGCUUAAUUUGUAAUAUUUUUUG -------.(..((((.((((((((.....)))))))).))))..)((((((......(((((....)))))))))))............ ( -15.40, z-score = -1.95, R) >droVir3.scaffold_12963 2071307 88 + 20206255 GAAAGCUGGUUGUAGCCUGUCAUUAAACAAAUGACGGCUGAUUUCCAAAUUUGUUU-CAGUUAUUUUGCUUAAUUUGUAAUAUUCAUUG (((...(((...(((((.((((((.....)))))))))))....))).........-........((((.......))))..))).... ( -18.00, z-score = -1.55, R) >droMoj3.scaffold_6500 27216539 88 + 32352404 CAAAGCGCAUUGUAGAUUGUCAUUAAACAAAUGACGGCUGAUUUCCAAAUUGGUUU-GAGUAAUUUUACUUAAUUUGUAAUAUUCCUUG ............(((.((((((((.....))))))))))).....(((((((((..-((....))..)).)))))))............ ( -13.30, z-score = -0.04, R) >droGri2.scaffold_15126 1095924 81 + 8399593 GAAAGCGCGUUGUAGCCUGUCAUUAAACAAAUGACGGCUGAUUUCCAAAUUUGUUUUGAGUAAUUUUACUUGUUUCGCAAU-------- ......(((...(((((.((((((.....))))))))))).................(((((....)))))....)))...-------- ( -18.40, z-score = -1.52, R) >consensus ____C_UUGUUGUGACCUGUCAUUAAACAAAUGACGGCUCAUUUCCAAAUUUGUACCGAGUAAUUUUGCUUAAUUUGUAAUAUUUUUUG ................((((((((.....))))))))........((((((......(((((....)))))))))))............ (-10.10 = -10.07 + -0.03)

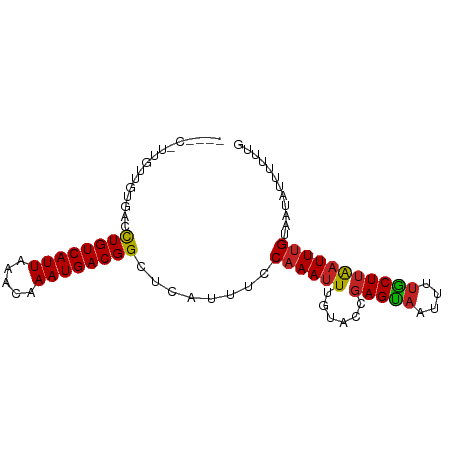
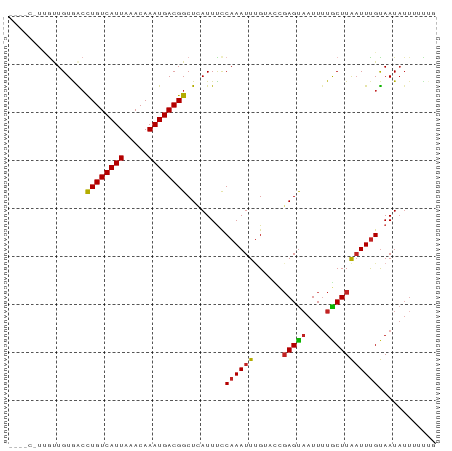
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:32:44 2011