| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,692,700 – 15,692,801 |
| Length | 101 |
| Max. P | 0.947674 |

| Location | 15,692,700 – 15,692,801 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 71.10 |
| Shannon entropy | 0.52179 |
| G+C content | 0.39728 |
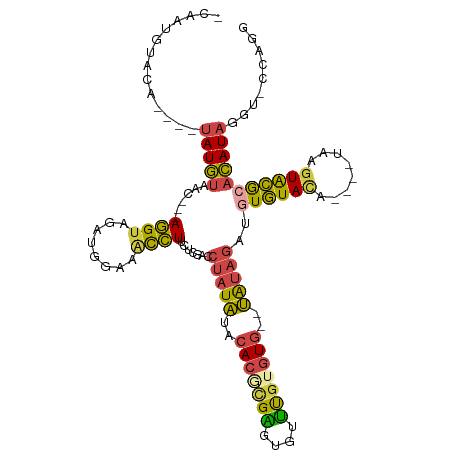
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -13.97 |
| Energy contribution | -13.28 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.56 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.947674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
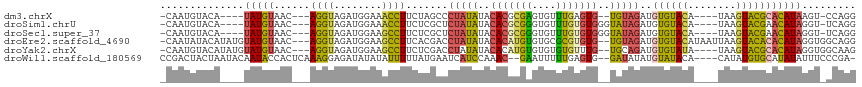
>dm3.chrX 15692700 101 - 22422827 -CAAUGUACA----UAUGUAAC---AGGUAGAUGGAAACCUUCUAGCCCUAUAUACACGCGAGUGUUUGAGUG--UGUAGAUGUGUACA----UAAGUACGCACAUAAGU-CCAGG -...((.((.----(((((...---.((((((.(....)..))).)))((((((((((....))))......)--)))))..((((((.----...))))))))))).))-.)).. ( -27.10, z-score = -1.31, R) >droSim1.chrU 270934 103 + 15797150 -CAAUGUACA----UAUGUAAC---AGGUAGAUGGAAACCUUCUCGCUCUAUAUACACGCGGGUGUUUGUGUGGGUAUAGAUGUGUACA----UAAGUACGAACAUAGGU-UCAGG -....((((.----((((((..---((((........))))...(((((((((..(((((((....)))))))..)))))).)))))))----)).))))((((....))-))... ( -32.00, z-score = -2.48, R) >droSec1.super_37 273469 103 - 454039 -CAAUGUACA----UAUGUAAC---AGGUAGAUGGAAGCCUUCUCGCUCUAUAUACACGCGGGUGUUUGUGUGGGUAUAGAUGUGUACA----UAAGUACGAACAUAGGU-UCAGG -....((((.----((((((..---((((........))))...(((((((((..(((((((....)))))))..)))))).)))))))----)).))))((((....))-))... ( -32.30, z-score = -2.40, R) >droEre2.scaffold_4690 7399318 110 + 18748788 -CAAUAUACAUAUGUAUGUAAC---AGGUAGAUGGAAGCCUUCACGACCUAUAUACACAUGUGUGCGCGUGUG--UGUAGAUGUGUACAUAAUUAAGUACACACAUAGGUGGCAGG -..((((((....))))))...---((((........))))...(.((((((((((((((((....)))))))--))))..(((((((........)))))))..))))).).... ( -35.10, z-score = -1.64, R) >droYak2.chrX 9892368 106 - 21770863 -CAAUGUACAUAUGUAUGUAAC---AGGUAGAUGGAAGCCUUCUCGACCUAUAUACACAUGUGUGUGUGUUUG--UGCAGAUGUGUAUA----UAAGUACGCACAUAGGUGGCAAG -...(((((.(((((.((((..---.(((.((.(((....))))).)))(((((((((((.((..(......)--..)).)))))))))----))..)))).))))).)).))).. ( -31.00, z-score = -0.80, R) >droWil1.scaffold_180569 424552 107 + 1405142 CCGACUACUAAUACAAUACCACUCAAAGGAGAUAUAUAUUUUUAUGAAUCAUCCAAAC--GAAUUUUUGAGUG--GAUAUAUGUAUACA----CAUAUGUGCAUAUAUUUCCCGA- .((...............(((((((((((((((.((((....)))).))).)))....--.....))))))))--)(((((((((((..----....)))))))))))....)).- ( -26.20, z-score = -4.00, R) >consensus _CAAUGUACA____UAUGUAAC___AGGUAGAUGGAAACCUUCUCGACCUAUAUACACGCGAGUGUUUGUGUG__UAUAGAUGUGUACA____UAAGUACGCACAUAGGU_CCAGG ..............(((((......((((........)))).......(((((..(((((((....)))))))..)))))..((((((........)))))))))))......... (-13.97 = -13.28 + -0.68)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:47:21 2011