| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,690,044 – 15,690,187 |
| Length | 143 |
| Max. P | 0.713857 |

| Location | 15,690,044 – 15,690,187 |
|---|---|
| Length | 143 |
| Sequences | 4 |
| Columns | 144 |
| Reading direction | forward |
| Mean pairwise identity | 64.05 |
| Shannon entropy | 0.57198 |
| G+C content | 0.46494 |
| Mean single sequence MFE | -33.60 |
| Consensus MFE | -10.16 |
| Energy contribution | -9.97 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.713857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
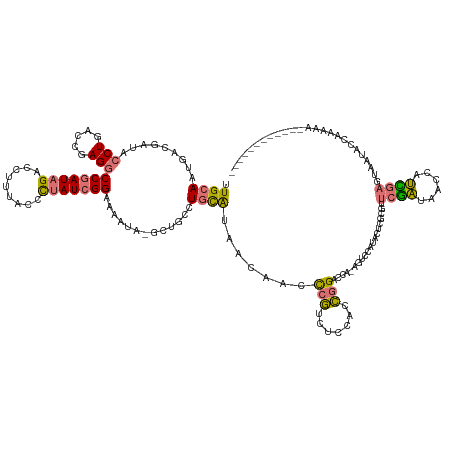
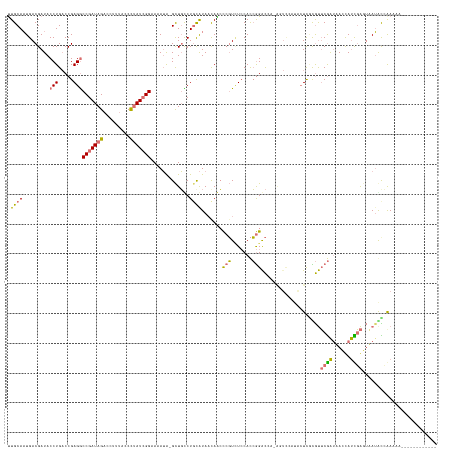
>dm3.chrX 15690044 143 + 22422827 CUGCACUGACGAUACCUGACCGAGGCCGAUAGACCUUUACCCUAUCGGUAAAUA-GCUGCCUGUAUAACAGCCCGUCUCCACCGGACCAAAGUCCACGCUCGCGUUCGAUAACUAUCGAGUUAGGUGAAAAAUCUCGGAUCUCA .....((((.((((((((((((((((((((((.........)))))))).....-((((.........))))...........((((....))))...))))...(((((....)))))))))))).....)))))))...... ( -40.00, z-score = -1.80, R) >droSec1.super_37 273997 128 + 454039 UUGCAAUGACGAUACCUGACCGAGGCCGAUAGACCUGGAACUCAUAGGGAAAAGUGUUUCCUGCGUA-CAGGUAUUAUACAUAAGGUUAUGUUUCAUACUCAUACGCAGGAAUCACUCGACCAUAUGAA--------------- ...................((.(((.(....).)))))...(((((.((...((((.((((((((((-..(((((...(((((....)))))...)))))..)))))))))).))))...)).))))).--------------- ( -40.40, z-score = -3.49, R) >droYak2.chrX 9892934 131 + 21770863 UUGCAAUGACGAUACCUGACCGAGGCCGAUAGACCUUUACCCUAUCGGAAAAUA-GCUGUCUCCAUAACAACCCGUUUCCACCGGACCAGAGUCCAUACUCCCGUUCGAUUAUCAUCGACUAAUACCGAAAA------------ ......(((.((((..(((.((.(((((((((.........)))))(((((...-(.(((.......))).)...)))))...)).)).((((....)))).)).)))..)))).)))..............------------ ( -24.70, z-score = -0.83, R) >droEre2.scaffold_4690 7399855 121 - 18748788 UUGCAAUGACGAUACCUGUCCGAGACCGAUAGACCUUUACCCUAUCGGAAAAUA-GCUGUCUGCAUAAUAACCCGUCUCCACCGGAC------------UUGUGUUCGAUUACUAUUGAGUAAUGCCAAUCACA---------- ..((.....((((((..(((((...(((((((.........)))))))......-((.....))..................)))))------------..))).)))((((((....))))))))........---------- ( -29.30, z-score = -2.72, R) >consensus UUGCAAUGACGAUACCUGACCGAGGCCGAUAGACCUUUACCCUAUCGGAAAAUA_GCUGCCUGCAUAACAACCCGUCUCCACCGGACCA_AGUCCAUACUCGCGUUCGAUAACCAUCGAGUAAUACCAAAAA____________ .((((.........(((.....)))(((((((.........))))))).............)))).......(((.......)))....................((((......))))......................... (-10.16 = -9.97 + -0.19)

| Location | 15,690,044 – 15,690,187 |
|---|---|
| Length | 143 |
| Sequences | 4 |
| Columns | 144 |
| Reading direction | reverse |
| Mean pairwise identity | 64.05 |
| Shannon entropy | 0.57198 |
| G+C content | 0.46494 |
| Mean single sequence MFE | -42.15 |
| Consensus MFE | -11.61 |
| Energy contribution | -11.42 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.623538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
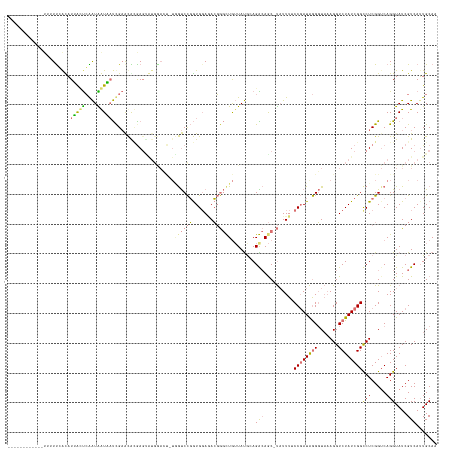
>dm3.chrX 15690044 143 - 22422827 UGAGAUCCGAGAUUUUUCACCUAACUCGAUAGUUAUCGAACGCGAGCGUGGACUUUGGUCCGGUGGAGACGGGCUGUUAUACAGGCAGC-UAUUUACCGAUAGGGUAAAGGUCUAUCGGCCUCGGUCAGGUAUCGUCAGUGCAG ...((.(((((..((((((((...((((...(((....))).))))...((((....))))))))))))..(((((((.....))))))-).....((((((((.......)))))))).)))))))..((((.....)))).. ( -48.70, z-score = -1.45, R) >droSec1.super_37 273997 128 - 454039 ---------------UUCAUAUGGUCGAGUGAUUCCUGCGUAUGAGUAUGAAACAUAACCUUAUGUAUAAUACCUG-UACGCAGGAAACACUUUUCCCUAUGAGUUCCAGGUCUAUCGGCCUCGGUCAGGUAUCGUCAUUGCAA ---------------((((((.((..(((((.(((((((((((..((((...(((((....)))))...))))..)-)))))))))).)))))..)).))))))..(((((((....))))).))....(((.......))).. ( -48.00, z-score = -5.42, R) >droYak2.chrX 9892934 131 - 21770863 ------------UUUUCGGUAUUAGUCGAUGAUAAUCGAACGGGAGUAUGGACUCUGGUCCGGUGGAAACGGGUUGUUAUGGAGACAGC-UAUUUUCCGAUAGGGUAAAGGUCUAUCGGCCUCGGUCAGGUAUCGUCAUUGCAA ------------...............(((((((...((.((((.((((((((..(..(((..((((((..(((((((.....))))))-)..))))))...)))..)..))))))..)))))).))...)))))))....... ( -40.60, z-score = -1.32, R) >droEre2.scaffold_4690 7399855 121 + 18748788 ----------UGUGAUUGGCAUUACUCAAUAGUAAUCGAACACAA------------GUCCGGUGGAGACGGGUUAUUAUGCAGACAGC-UAUUUUCCGAUAGGGUAAAGGUCUAUCGGUCUCGGACAGGUAUCGUCAUUGCAA ----------((..((.(((((((((....)))))).((..((..------------((((....((((((((..((...((.....))-.))..)))((((((.......)))))).)))))))))..)).)))))))..)). ( -31.30, z-score = -0.74, R) >consensus ____________UUUUUCACAUUACUCAAUAAUAAUCGAACACGAGUAUGGACU_UGGUCCGGUGGAGACGGGCUGUUAUGCAGACAGC_UAUUUUCCGAUAGGGUAAAGGUCUAUCGGCCUCGGUCAGGUAUCGUCAUUGCAA .....................(((((....))))).....................((((((.......))))))........(((..........((((((((.......))))))))(((.....)))....)))....... (-11.61 = -11.42 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:47:20 2011