| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,680,632 – 15,680,790 |
| Length | 158 |
| Max. P | 0.993669 |

| Location | 15,680,632 – 15,680,750 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.61 |
| Shannon entropy | 0.25027 |
| G+C content | 0.51359 |
| Mean single sequence MFE | -38.86 |
| Consensus MFE | -28.04 |
| Energy contribution | -30.12 |
| Covariance contribution | 2.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.869243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15680632 118 + 22422827 UGCCCUCGAGUUUGCCCACGACUACUACAAGUGGGUACA--ACGAGGUCCGCUUUCAACGAACCAAGUUCUCCGGCGAGUUUGUGGACAGUAGUUGGUCCAACUCACUUAAGAACUCGGA .(((((((....(((((((...........)))))))..--.)))))...))..........((.((((((..((.((((...(((((........))))))))).))..)))))).)). ( -40.20, z-score = -1.80, R) >droSim1.chrX 12162514 118 + 17042790 UGCCCUCGAGUUUGCCCACGACUACUACAAGUGGGUACA--ACGAGGUCCGCUUCCAACGAACCAAGUUCUCCGGCGAGUUUGCGGACAGUCGCUUGUCCAACUCACCUAAGAACUCGGA .(((((((....(((((((...........)))))))..--.)))))...))..........((.((((((..((.(((((...((((((....))))))))))).))..)))))).)). ( -43.00, z-score = -2.81, R) >droSec1.super_37 264723 118 + 454039 UGCCCUCGAGUUUGCCCACGACUACUACAAGUGGGUACA--ACGAGGUCGUCUUCCAACGAACCAAGUUCUCCGGCGAGUUUGCGGACAGUAGCUUGUCCAACUCACUUAAGAACUCGGG ...(((((....(((((((...........)))))))..--.)))))((((......)))).((.((((((..((.(((((...((((((....))))))))))).))..)))))).)). ( -43.10, z-score = -2.81, R) >droYak2.chrX 9883966 120 + 21770863 UGCCCUCGAAUUCGCCCACGACUACUACAAGUGGGUAAAAAAUGAGGCCCACUCUCAUCGAACCGAGUUCUCAUGCGUGUUGGUGGGCAGAAGCUUCCCAAACUCACUUAAGAACUCUAA .....((((....((((((...........))))))......(((((.....)))))))))...(((((((...(.(..((((..(((....)))..))))...).)...)))))))... ( -35.60, z-score = -2.43, R) >droEre2.scaffold_4690 7390560 118 - 18748788 UGCCCUUGAGUUUGCACACGACUACUACAAGUGGGUAAA--AUGAGGCCCCCUUUCAACAAACCGAGUUCUCAUGCGAGUUUGUGGGCAGUGACUUCGCAAACUCACUUAAGAACUCUGA ...((((...(((((.(((...........))).)))))--..)))).................(((((((...(.(((((((((((.......))))))))))).)...)))))))... ( -32.40, z-score = -0.95, R) >consensus UGCCCUCGAGUUUGCCCACGACUACUACAAGUGGGUACA__ACGAGGUCCGCUUUCAACGAACCAAGUUCUCCGGCGAGUUUGUGGACAGUAGCUUGUCCAACUCACUUAAGAACUCGGA ...(((((....(((((((...........))))))).....)))))...............((.((((((..((.(((((...((((((....))))))))))).))..)))))).)). (-28.04 = -30.12 + 2.08)


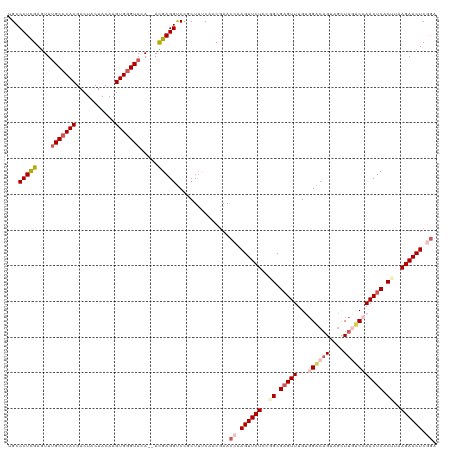
| Location | 15,680,632 – 15,680,750 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.61 |
| Shannon entropy | 0.25027 |
| G+C content | 0.51359 |
| Mean single sequence MFE | -44.02 |
| Consensus MFE | -33.22 |
| Energy contribution | -34.94 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.939450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15680632 118 - 22422827 UCCGAGUUCUUAAGUGAGUUGGACCAACUACUGUCCACAAACUCGCCGGAGAACUUGGUUCGUUGAAAGCGGACCUCGU--UGUACCCACUUGUAGUAGUCGUGGGCAAACUCGAGGGCA .((((((((((..(((((((((((........)))))...))))))..))))))))))(((((.....)))))(((((.--.((.(((((...........)))))...)).)))))... ( -46.60, z-score = -3.04, R) >droSim1.chrX 12162514 118 - 17042790 UCCGAGUUCUUAGGUGAGUUGGACAAGCGACUGUCCGCAAACUCGCCGGAGAACUUGGUUCGUUGGAAGCGGACCUCGU--UGUACCCACUUGUAGUAGUCGUGGGCAAACUCGAGGGCA .((((((((((.(((((((((((((......)))))...)))))))).))))))))))(((((.....)))))(((((.--.((.(((((...........)))))...)).)))))... ( -51.00, z-score = -3.15, R) >droSec1.super_37 264723 118 - 454039 CCCGAGUUCUUAAGUGAGUUGGACAAGCUACUGUCCGCAAACUCGCCGGAGAACUUGGUUCGUUGGAAGACGACCUCGU--UGUACCCACUUGUAGUAGUCGUGGGCAAACUCGAGGGCA .((((((((((..((((((((((((......)))))...)))))))..)))))))))).(((((....)))))(((((.--.((.(((((...........)))))...)).)))))... ( -48.90, z-score = -3.29, R) >droYak2.chrX 9883966 120 - 21770863 UUAGAGUUCUUAAGUGAGUUUGGGAAGCUUCUGCCCACCAACACGCAUGAGAACUCGGUUCGAUGAGAGUGGGCCUCAUUUUUUACCCACUUGUAGUAGUCGUGGGCGAAUUCGAGGGCA ...(((((((((.(((.(((.((...((....))...))))).))).)))))))))...............(.((((....((..(((((...........)))))..))...)))).). ( -38.70, z-score = -1.25, R) >droEre2.scaffold_4690 7390560 118 + 18748788 UCAGAGUUCUUAAGUGAGUUUGCGAAGUCACUGCCCACAAACUCGCAUGAGAACUCGGUUUGUUGAAAGGGGGCCUCAU--UUUACCCACUUGUAGUAGUCGUGUGCAAACUCAAGGGCA ...(((((((((.(((((((((.(...........).))))))))).)))))))))(((((((...((((((.......--....))).))).............)))))))........ ( -34.89, z-score = -0.77, R) >consensus UCCGAGUUCUUAAGUGAGUUGGACAAGCUACUGUCCACAAACUCGCCGGAGAACUUGGUUCGUUGAAAGCGGACCUCGU__UGUACCCACUUGUAGUAGUCGUGGGCAAACUCGAGGGCA .((((((((((..((((((((((((......))))))...))))))..)))))))))).............(.(((((.......(((((...........)))))......))))).). (-33.22 = -34.94 + 1.72)

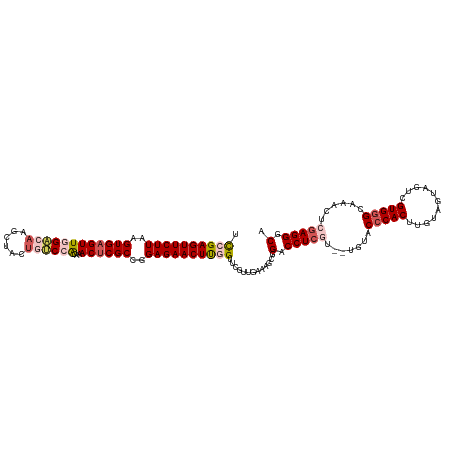
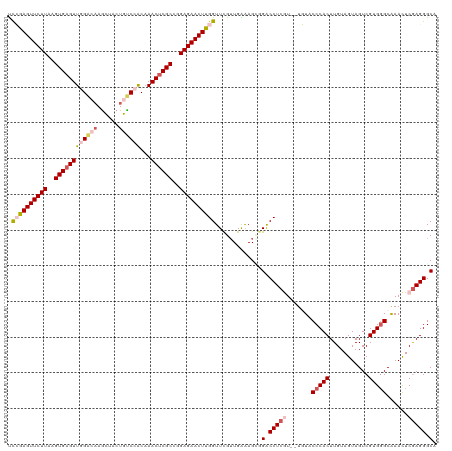
| Location | 15,680,670 – 15,680,790 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.83 |
| Shannon entropy | 0.28641 |
| G+C content | 0.50833 |
| Mean single sequence MFE | -42.96 |
| Consensus MFE | -27.60 |
| Energy contribution | -29.80 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.740212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15680670 120 + 22422827 AACGAGGUCCGCUUUCAACGAACCAAGUUCUCCGGCGAGUUUGUGGACAGUAGUUGGUCCAACUCACUUAAGAACUCGGAUUGGUUGGAGUGGUCCCAGUUACAAGUUCGACCGUCUGAU (((..((.((((((.((((.((((.((((((..((.((((...(((((........))))))))).))..)))))).)).)).)))))))))).))..)))................... ( -41.30, z-score = -1.74, R) >droSim1.chrX 12162552 120 + 17042790 AACGAGGUCCGCUUCCAACGAACCAAGUUCUCCGGCGAGUUUGCGGACAGUCGCUUGUCCAACUCACCUAAGAACUCGGAUUGGUUGGAGUGGCCCUAGUUACAAGUUGGGUCGUCUGAU (((.(((.(((((.(((((.((((.((((((..((.(((((...((((((....))))))))))).))..)))))).)).)).)))))))))).))).)))....((..(.....)..)) ( -47.40, z-score = -2.79, R) >droSec1.super_37 264761 120 + 454039 AACGAGGUCGUCUUCCAACGAACCAAGUUCUCCGGCGAGUUUGCGGACAGUAGCUUGUCCAACUCACUUAAGAACUCGGGUUGGUUGGAGUGGCCCUAGUUACAAGUUCGGUCGUCUGAU (((..(((((.((.(((((.((((.((((((..((.(((((...((((((....))))))))))).))..))))))..)))).))))))))))))...)))......((((....)))). ( -44.90, z-score = -2.48, R) >droYak2.chrX 9884006 120 + 21770863 AAUGAGGCCCACUCUCAUCGAACCGAGUUCUCAUGCGUGUUGGUGGGCAGAAGCUUCCCAAACUCACUUAAGAACUCUAAUUGGUUGAAGUGGGCCGAGAUACAAGUUCGGUGGUCUGGU .....((((((((.(((((((...(((((((...(.(..((((..(((....)))..))))...).)...)))))))...)))).))))))))))).((((((.......)).))))... ( -41.60, z-score = -1.81, R) >droEre2.scaffold_4690 7390598 120 - 18748788 AAUGAGGCCCCCUUUCAACAAACCGAGUUCUCAUGCGAGUUUGUGGGCAGUGACUUCGCAAACUCACUUAAGAACUCUGAUUGGUUGGAGUGGCUCGAGUUACAAGUACGGUCGUCUGGU .((.((((..((.((((((...(.(((((((...(.(((((((((((.......))))))))))).)...))))))).)....))))))(((((....)))))......))..)))).)) ( -39.60, z-score = -1.49, R) >consensus AACGAGGUCCGCUUUCAACGAACCAAGUUCUCCGGCGAGUUUGUGGACAGUAGCUUGUCCAACUCACUUAAGAACUCGGAUUGGUUGGAGUGGCCCGAGUUACAAGUUCGGUCGUCUGAU (((..((.(((((.(((((.((((.((((((..((.(((((...((((((....))))))))))).))..)))))).)).)).)))))))))).))..)))................... (-27.60 = -29.80 + 2.20)


| Location | 15,680,670 – 15,680,790 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.83 |
| Shannon entropy | 0.28641 |
| G+C content | 0.50833 |
| Mean single sequence MFE | -43.14 |
| Consensus MFE | -30.28 |
| Energy contribution | -31.32 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.993669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15680670 120 - 22422827 AUCAGACGGUCGAACUUGUAACUGGGACCACUCCAACCAAUCCGAGUUCUUAAGUGAGUUGGACCAACUACUGUCCACAAACUCGCCGGAGAACUUGGUUCGUUGAAAGCGGACCUCGUU ....((.((((...(((.((((((((.....))))......((((((((((..(((((((((((........)))))...))))))..))))))))))...)))).)))..))))))... ( -41.10, z-score = -2.27, R) >droSim1.chrX 12162552 120 - 17042790 AUCAGACGACCCAACUUGUAACUAGGGCCACUCCAACCAAUCCGAGUUCUUAGGUGAGUUGGACAAGCGACUGUCCGCAAACUCGCCGGAGAACUUGGUUCGUUGGAAGCGGACCUCGUU ....((((((((............)))((.(((((((.((.((((((((((.(((((((((((((......)))))...)))))))).)))))))))))).))))).)).))...))))) ( -50.40, z-score = -4.19, R) >droSec1.super_37 264761 120 - 454039 AUCAGACGACCGAACUUGUAACUAGGGCCACUCCAACCAACCCGAGUUCUUAAGUGAGUUGGACAAGCUACUGUCCGCAAACUCGCCGGAGAACUUGGUUCGUUGGAAGACGACCUCGUU ....(((((..(..((((....))))..)..((((((.((((.((((((((..((((((((((((......)))))...)))))))..)))))))))))).))))))........))))) ( -43.30, z-score = -3.06, R) >droYak2.chrX 9884006 120 - 21770863 ACCAGACCACCGAACUUGUAUCUCGGCCCACUUCAACCAAUUAGAGUUCUUAAGUGAGUUUGGGAAGCUUCUGCCCACCAACACGCAUGAGAACUCGGUUCGAUGAGAGUGGGCCUCAUU ........................(((((((((((.(.((((.(((((((((.(((.(((.((...((....))...))))).))).))))))))))))).).))).))))))))..... ( -41.40, z-score = -3.24, R) >droEre2.scaffold_4690 7390598 120 + 18748788 ACCAGACGACCGUACUUGUAACUCGAGCCACUCCAACCAAUCAGAGUUCUUAAGUGAGUUUGCGAAGUCACUGCCCACAAACUCGCAUGAGAACUCGGUUUGUUGAAAGGGGGCCUCAUU ((.((((....)).)).)).....(((((.(((((((.((((.(((((((((.(((((((((.(...........).))))))))).))))))))))))).))))...))))).)))... ( -39.50, z-score = -2.72, R) >consensus AUCAGACGACCGAACUUGUAACUAGGGCCACUCCAACCAAUCCGAGUUCUUAAGUGAGUUGGACAAGCUACUGUCCACAAACUCGCCGGAGAACUUGGUUCGUUGAAAGCGGACCUCGUU ........................((.((.(((((((.((((.((((((((..((((((((((((......))))))...))))))..)))))))))))).)))))).).)).))..... (-30.28 = -31.32 + 1.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:47:17 2011