
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,656,716 – 15,656,857 |
| Length | 141 |
| Max. P | 0.979406 |

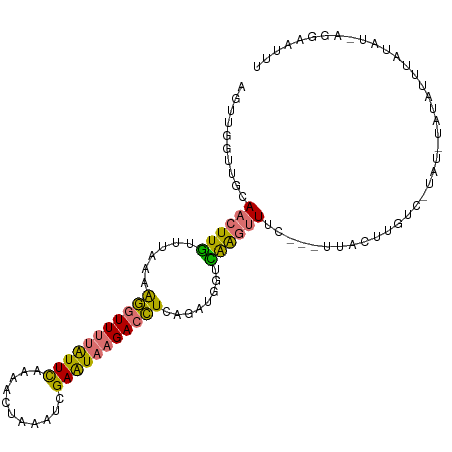
| Location | 15,656,716 – 15,656,819 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 65.62 |
| Shannon entropy | 0.56191 |
| G+C content | 0.27719 |
| Mean single sequence MFE | -18.53 |
| Consensus MFE | -9.85 |
| Energy contribution | -9.17 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.661663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15656716 103 + 22422827 AGUUGGUUGUAACUUAUUUAAGAGGUUUUAUUCAAACCUAAAUCGAAUAAGACCUCAGAUGGUUAGGUUUC---UUAUUUGUUUCAU-UAUAUUCAUAU-AGGAAUUU .......(.(((((.((((..((((((((((((...........))))))))))))))))))))).).(((---((((.((......-......)).))-)))))... ( -21.60, z-score = -1.86, R) >droSim1.chrX 12139000 96 + 17042790 AGUUGGUUGCAACUUGUUUAAAAGGUUUUAUUCAAAACUAAAUCGAAUAAGACCUCAGAUGGUCAAGUUUC---UUAUUU--------UAUAUUUAUGU-AGGAAUUU ..........((((((......(((((((((((...........)))))))))))........))))))((---((((..--------.........))-)))).... ( -17.74, z-score = -1.28, R) >droSec1.super_37 240941 105 + 454039 AGUUGGUUGCAACUUGUUUAAAAGGUUUUAUUCAAAACUAAAUCGAAUAAGACCUCAGAUGGUCAAGUUUC---UUACUCGUCCUAUUUAUAUUUUUUUUAGGAAUUU ....(((.(.((((((......(((((((((((...........)))))))))))........)))))).)---..)))..(((((.............))))).... ( -20.26, z-score = -1.57, R) >triCas2.ChLG5 9533398 107 + 18847211 AAUAAAGAUUAAGUUGAU-ACAGAAUUUUGUUUUAAUUGCCUCCGAGUCUGAGUUCUGAUACUUGAACUAUAAAUUACUUAUCGUCGAUUUUUUUAUAUCUUGGGUUU ....(((((.((((((((-.((((.....((.......)).......))))(((((........)))))..............))))))))......)))))...... ( -14.50, z-score = 0.73, R) >consensus AGUUGGUUGCAACUUGUUUAAAAGGUUUUAUUCAAAACUAAAUCGAAUAAGACCUCAGAUGGUCAAGUUUC___UUACUUGUC_UAU_UAUAUUUAUAU_AGGAAUUU ..........((((((......(((((((((((...........)))))))))))........))))))....................................... ( -9.85 = -9.17 + -0.69)


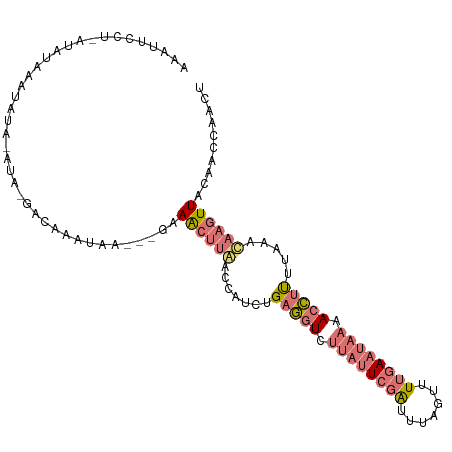
| Location | 15,656,716 – 15,656,819 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 65.62 |
| Shannon entropy | 0.56191 |
| G+C content | 0.27719 |
| Mean single sequence MFE | -15.80 |
| Consensus MFE | -7.69 |
| Energy contribution | -9.50 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.949114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15656716 103 - 22422827 AAAUUCCU-AUAUGAAUAUA-AUGAAACAAAUAA---GAAACCUAACCAUCUGAGGUCUUAUUCGAUUUAGGUUUGAAUAAAACCUCUUAAAUAAGUUACAACCAACU ..((((..-....))))...-.((.(((.....(---((..........)))(((((.((((((((.......)))))))).)))))........))).))....... ( -15.10, z-score = -1.62, R) >droSim1.chrX 12139000 96 - 17042790 AAAUUCCU-ACAUAAAUAUA--------AAAUAA---GAAACUUGACCAUCUGAGGUCUUAUUCGAUUUAGUUUUGAAUAAAACCUUUUAAACAAGUUGCAACCAACU ........-...........--------......---(.((((((.......(((((.((((((((.......)))))))).))))).....)))))).)........ ( -17.00, z-score = -2.98, R) >droSec1.super_37 240941 105 - 454039 AAAUUCCUAAAAAAAAUAUAAAUAGGACGAGUAA---GAAACUUGACCAUCUGAGGUCUUAUUCGAUUUAGUUUUGAAUAAAACCUUUUAAACAAGUUGCAACCAACU ....(((((.............)))))(((((..---...))))).......(((((.((((((((.......)))))))).))))).......(((((....))))) ( -20.02, z-score = -2.55, R) >triCas2.ChLG5 9533398 107 - 18847211 AAACCCAAGAUAUAAAAAAAUCGACGAUAAGUAAUUUAUAGUUCAAGUAUCAGAACUCAGACUCGGAGGCAAUUAAAACAAAAUUCUGU-AUCAACUUAAUCUUUAUU ........(((........)))...(((((((.......(((((........)))))..((..(((((...............))))).-.)).)))).)))...... ( -11.06, z-score = 0.08, R) >consensus AAAUUCCU_AUAUAAAUAUA_AUA_GACAAAUAA___GAAACUUAACCAUCUGAGGUCUUAUUCGAUUUAGUUUUGAAUAAAACCUUUUAAACAAGUUACAACCAACU .......................................((((((.......(((((.((((((((.......)))))))).))))).....)))))).......... ( -7.69 = -9.50 + 1.81)


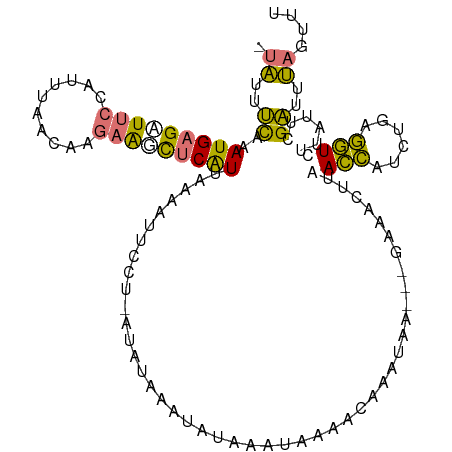
| Location | 15,656,750 – 15,656,857 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 64.66 |
| Shannon entropy | 0.57854 |
| G+C content | 0.29752 |
| Mean single sequence MFE | -21.80 |
| Consensus MFE | -13.10 |
| Energy contribution | -13.22 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.979406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15656750 107 + 22422827 AACCUAAAUCGAAUAAGACCUCAGAUGGUUAGGUUUC---UUAUUUGUUUCAUU-AUAUUCAUAU-AGGAAUUUUAAUGAGCUUCUUUUUAAGGGGAAGCUCAUUUAAAAUA- (((((((.((......))((......)))))))))((---((((.((.......-.....)).))-))))((((((((((((((((((....))))))))))).))))))).- ( -27.40, z-score = -3.23, R) >droSim1.chrX 12139034 100 + 17042790 AAACUAAAUCGAAUAAGACCUCAGAUGGUCAAGUUUC---UUAUU--UUAUAUUUAUGU-------AGGAAUUUUGACGAGCUUCUUGUUAAAUGGAAUCUCAUUUGAAAUA- ....................((((((.((((((.(((---((((.--..........))-------))))).))))))(((.((((........)))).)))))))))....- ( -20.90, z-score = -1.69, R) >droSec1.super_37 240975 109 + 454039 AAACUAAAUCGAAUAAGACCUCAGAUGGUCAAGUUUC---UUACUCGUCCUAUUUAUAUUUUUUUUAGGAAUUUUAAUGAAAUUCUUGUUAAAUGGAAUCUCAUUUGAACUA- ...(((((..(((((.((((......)))).(((...---..)))...........)))))..)))))...(((.(((((.(((((........))))).))))).)))...- ( -19.00, z-score = -1.01, R) >triCas2.ChLG5 9533431 110 + 18847211 AAUUGCCUCCGAGUCUGAGUUCUGAUACUUGAACUAUAAAUUACUUAUCGUCGAUUUUUUUAUAUCUUGGGUUUUUAUGCGACUGCUGUUA--UGGCA-CUCAUUUGGGUGAA ..(..((((((((....(((((........)))))..(((((((.....)).)))))........)))))..........((.((((....--.))))-.))....)))..). ( -19.90, z-score = -0.13, R) >consensus AAACUAAAUCGAAUAAGACCUCAGAUGGUCAAGUUUC___UUACUUGUCAUAUUUAUAUUUAUAU_AGGAAUUUUAAUGAGAUUCUUGUUAAAUGGAAUCUCAUUUGAAAUA_ ........((((((..((((......))))................................................((((((((........))))))))))))))..... (-13.10 = -13.22 + 0.12)



| Location | 15,656,750 – 15,656,857 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 64.66 |
| Shannon entropy | 0.57854 |
| G+C content | 0.29752 |
| Mean single sequence MFE | -17.18 |
| Consensus MFE | -7.72 |
| Energy contribution | -7.35 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.757007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15656750 107 - 22422827 -UAUUUUAAAUGAGCUUCCCCUUAAAAAGAAGCUCAUUAAAAUUCCU-AUAUGAAUAU-AAUGAAACAAAUAA---GAAACCUAACCAUCUGAGGUCUUAUUCGAUUUAGGUU -..(((((((((((((((..........))))))))))...((((..-....))))..-...........)))---))(((((((..(((.(((......))))))))))))) ( -20.70, z-score = -2.75, R) >droSim1.chrX 12139034 100 - 17042790 -UAUUUCAAAUGAGAUUCCAUUUAACAAGAAGCUCGUCAAAAUUCCU-------ACAUAAAUAUAA--AAUAA---GAAACUUGACCAUCUGAGGUCUUAUUCGAUUUAGUUU -((..((.((((((((..((........(.....)(((((..(((..-------............--.....---)))..)))))....))..)))))))).))..)).... ( -12.81, z-score = -0.25, R) >droSec1.super_37 240975 109 - 454039 -UAGUUCAAAUGAGAUUCCAUUUAACAAGAAUUUCAUUAAAAUUCCUAAAAAAAAUAUAAAUAGGACGAGUAA---GAAACUUGACCAUCUGAGGUCUUAUUCGAUUUAGUUU -.......((((((((((..........)))))))))).....(((((.............)))))(((((((---((..(((........))).)))))))))......... ( -21.72, z-score = -2.31, R) >triCas2.ChLG5 9533431 110 - 18847211 UUCACCCAAAUGAG-UGCCA--UAACAGCAGUCGCAUAAAAACCCAAGAUAUAAAAAAAUCGACGAUAAGUAAUUUAUAGUUCAAGUAUCAGAACUCAGACUCGGAGGCAAUU ..............-((((.--.....((....))........((..(((........))).................(((((........))))).......)).))))... ( -13.50, z-score = -0.27, R) >consensus _UAUUUCAAAUGAGAUUCCAUUUAACAAGAAGCUCAUUAAAAUUCCU_AUAUAAAUAUAAAUAAAACAAAUAA___GAAACUUAACCAUCUGAGGUCUUAUUCGAUUUAGUUU .((..((..(((((((((..........)))))))))..............................................((((......))))......))..)).... ( -7.72 = -7.35 + -0.37)

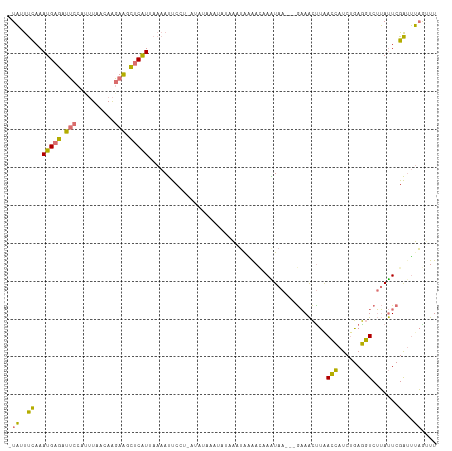
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:47:14 2011