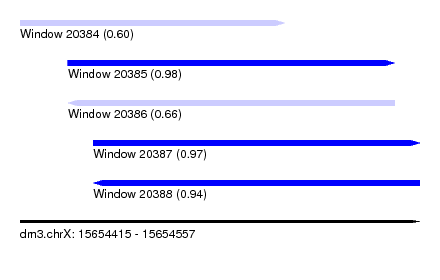
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,654,415 – 15,654,557 |
| Length | 142 |
| Max. P | 0.978114 |

| Location | 15,654,415 – 15,654,509 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Shannon entropy | -0.00000 |
| G+C content | 0.45745 |
| Mean single sequence MFE | -21.40 |
| Consensus MFE | -21.40 |
| Energy contribution | -21.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.95 |
| Structure conservation index | 1.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.596133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15654415 94 + 22422827 CUCUUAGUUACGAAAGGCAACCGUCGGUAGUUUUUCGCGUACCGCAACCGCACACACAUUUGUGCCCGUGUUAAAAUUACUCACAGAGUAAACA ((((..(((.(....)..))).((.(((((((((.((((....((....)).((((....))))..))))..))))))))).))))))...... ( -21.40, z-score = -0.95, R) >droSim1.chrX 12136694 94 + 17042790 CUCUUAGUUACGAAAGGCAACCGUCGGUAGUUUUUCGCGUACCGCAACCGCACACACAUUUGUGCCCGUGUUAAAAUUACUCACAGAGUAAACA ((((..(((.(....)..))).((.(((((((((.((((....((....)).((((....))))..))))..))))))))).))))))...... ( -21.40, z-score = -0.95, R) >droSec1.super_37 238628 94 + 454039 CUCUUAGUUACGAAAGGCAACCGUCGGUAGUUUUUCGCGUACCGCAACCGCACACACAUUUGUGCCCGUGUUAAAAUUACUCACAGAGUAAACA ((((..(((.(....)..))).((.(((((((((.((((....((....)).((((....))))..))))..))))))))).))))))...... ( -21.40, z-score = -0.95, R) >droYak2.chrX 9859442 94 + 21770863 CUCUUAGUUACGAAAGGCAACCGUCGGUAGUUUUUCGCGUACCGCAACCGCACACACAUUUGUGCCCGUGUUAAAAUUACUCACAGAGUAAACA ((((..(((.(....)..))).((.(((((((((.((((....((....)).((((....))))..))))..))))))))).))))))...... ( -21.40, z-score = -0.95, R) >droEre2.scaffold_4690 7364844 94 - 18748788 CUCUUAGUUACGAAAGGCAACCGUCGGUAGUUUUUCGCGUACCGCAACCGCACACACAUUUGUGCCCGUGUUAAAAUUACUCACAGAGUAAACA ((((..(((.(....)..))).((.(((((((((.((((....((....)).((((....))))..))))..))))))))).))))))...... ( -21.40, z-score = -0.95, R) >consensus CUCUUAGUUACGAAAGGCAACCGUCGGUAGUUUUUCGCGUACCGCAACCGCACACACAUUUGUGCCCGUGUUAAAAUUACUCACAGAGUAAACA ((((..(((.(....)..))).((.(((((((((.((((....((....)).((((....))))..))))..))))))))).))))))...... (-21.40 = -21.40 + 0.00)

| Location | 15,654,432 – 15,654,548 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.48 |
| Shannon entropy | 0.39422 |
| G+C content | 0.39148 |
| Mean single sequence MFE | -26.44 |
| Consensus MFE | -15.23 |
| Energy contribution | -19.40 |
| Covariance contribution | 4.17 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.978114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15654432 116 + 22422827 CAACCGUCGGUAGUUUUUCGCGU---ACCGCAACCGCACACACAUUUGUGCCCGUGUUAAAAUUACUCACAGAGUAAACAUUAGUG-UAUGAGUAAUUUCCCAAUAUCUAAUGUUGCUCU .....((.(((((((((.((((.---...((....)).((((....))))..))))..))))))))).))(((((((.((((((((-(..(((....)))...))).))))))))))))) ( -29.60, z-score = -3.11, R) >droSim1.chrX 12136711 116 + 17042790 CAACCGUCGGUAGUUUUUCGCGU---ACCGCAACCGCACACACAUUUGUGCCCGUGUUAAAAUUACUCACAGAGUAAACAUUAGUG-UAUGAGUAAUUUCCCAAUAUCUAAUGUUGCUCU .....((.(((((((((.((((.---...((....)).((((....))))..))))..))))))))).))(((((((.((((((((-(..(((....)))...))).))))))))))))) ( -29.60, z-score = -3.11, R) >droSec1.super_37 238645 116 + 454039 CAACCGUCGGUAGUUUUUCGCGU---ACCGCAACCGCACACACAUUUGUGCCCGUGUUAAAAUUACUCACAGAGUAAACAUUAGUG-UAUGAGUAAUUUCCCAAUAUCUAAUGUCGCUCU .....((.(((((((((.((((.---...((....)).((((....))))..))))..))))))))).))(((((..(((((((((-(..(((....)))...))).))))))).))))) ( -27.70, z-score = -2.65, R) >droYak2.chrX 9859459 117 + 21770863 CAACCGUCGGUAGUUUUUCGCGU---ACCGCAACCGCACACACAUUUGUGCCCGUGUUAAAAUUACUCACAGAGUAAACAUUAGUGGUAUGAGUAAUUUCCCAACAUCUAAUGUUGCUCU .....((.(((((((((.((((.---...((....)).((((....))))..))))..))))))))).))(((((((.((((((((....(((....)))....)).))))))))))))) ( -31.10, z-score = -2.90, R) >droEre2.scaffold_4690 7364861 116 - 18748788 CAACCGUCGGUAGUUUUUCGCGU---ACCGCAACCGCACACACAUUUGUGCCCGUGUUAAAAUUACUCACAGAGUAAACAUUAGUG-UAUGAGUAAUUUCCCAAUAUCUAAUGUUGCUCU .....((.(((((((((.((((.---...((....)).((((....))))..))))..))))))))).))(((((((.((((((((-(..(((....)))...))).))))))))))))) ( -29.60, z-score = -3.11, R) >droWil1.scaffold_181150 2480663 97 + 4952429 AAACUGUUUGCAUUUUUUACAGUCAAAAAGCAAACGAACAAAAAUUUACAAA-AUAUUAAUAUAAACGAUA--UUACAUAAGAACAUUUUUGGUAGCGCC-------------------- ....(((((((.(((((.......))))))))))))..(((((((.......-....((((((.....)))--))).........)))))))........-------------------- ( -11.05, z-score = -0.27, R) >consensus CAACCGUCGGUAGUUUUUCGCGU___ACCGCAACCGCACACACAUUUGUGCCCGUGUUAAAAUUACUCACAGAGUAAACAUUAGUG_UAUGAGUAAUUUCCCAAUAUCUAAUGUUGCUCU .....((.(((((((((..(((......)))....(((((......))))).......))))))))).))(((((((.((((((......(((....))).......))))))))))))) (-15.23 = -19.40 + 4.17)

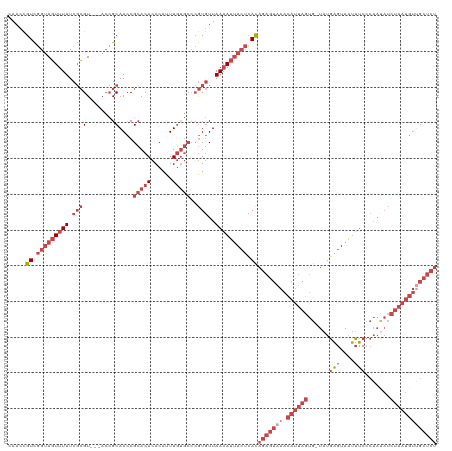
| Location | 15,654,432 – 15,654,548 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.48 |
| Shannon entropy | 0.39422 |
| G+C content | 0.39148 |
| Mean single sequence MFE | -27.92 |
| Consensus MFE | -19.70 |
| Energy contribution | -21.07 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.659322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15654432 116 - 22422827 AGAGCAACAUUAGAUAUUGGGAAAUUACUCAUA-CACUAAUGUUUACUCUGUGAGUAAUUUUAACACGGGCACAAAUGUGUGUGCGGUUGCGGU---ACGCGAAAAACUACCGACGGUUG ...(((((............(((((((((((((-...............)))))))))))))....((.(((((....))))).)))))))(((---(..........))))........ ( -30.36, z-score = -1.43, R) >droSim1.chrX 12136711 116 - 17042790 AGAGCAACAUUAGAUAUUGGGAAAUUACUCAUA-CACUAAUGUUUACUCUGUGAGUAAUUUUAACACGGGCACAAAUGUGUGUGCGGUUGCGGU---ACGCGAAAAACUACCGACGGUUG ...(((((............(((((((((((((-...............)))))))))))))....((.(((((....))))).)))))))(((---(..........))))........ ( -30.36, z-score = -1.43, R) >droSec1.super_37 238645 116 - 454039 AGAGCGACAUUAGAUAUUGGGAAAUUACUCAUA-CACUAAUGUUUACUCUGUGAGUAAUUUUAACACGGGCACAAAUGUGUGUGCGGUUGCGGU---ACGCGAAAAACUACCGACGGUUG ...(((((............(((((((((((((-...............)))))))))))))....((.(((((....))))).)))))))(((---(..........))))........ ( -30.06, z-score = -1.20, R) >droYak2.chrX 9859459 117 - 21770863 AGAGCAACAUUAGAUGUUGGGAAAUUACUCAUACCACUAAUGUUUACUCUGUGAGUAAUUUUAACACGGGCACAAAUGUGUGUGCGGUUGCGGU---ACGCGAAAAACUACCGACGGUUG ...(((((......((((..(((((((((((((................)))))))))))))))))((.(((((....))))).)))))))(((---(..........))))........ ( -31.39, z-score = -1.09, R) >droEre2.scaffold_4690 7364861 116 + 18748788 AGAGCAACAUUAGAUAUUGGGAAAUUACUCAUA-CACUAAUGUUUACUCUGUGAGUAAUUUUAACACGGGCACAAAUGUGUGUGCGGUUGCGGU---ACGCGAAAAACUACCGACGGUUG ...(((((............(((((((((((((-...............)))))))))))))....((.(((((....))))).)))))))(((---(..........))))........ ( -30.36, z-score = -1.43, R) >droWil1.scaffold_181150 2480663 97 - 4952429 --------------------GGCGCUACCAAAAAUGUUCUUAUGUAA--UAUCGUUUAUAUUAAUAU-UUUGUAAAUUUUUGUUCGUUUGCUUUUUGACUGUAAAAAAUGCAAACAGUUU --------------------((.....)).((((((((...((((((--......)))))).)))))-)))..............(((((((((((.......))))).))))))..... ( -15.00, z-score = -0.63, R) >consensus AGAGCAACAUUAGAUAUUGGGAAAUUACUCAUA_CACUAAUGUUUACUCUGUGAGUAAUUUUAACACGGGCACAAAUGUGUGUGCGGUUGCGGU___ACGCGAAAAACUACCGACGGUUG ....((((............(((((((((((((..((....))......)))))))))))))....((((((((......)))))..(((((......))))).......)))...)))) (-19.70 = -21.07 + 1.37)

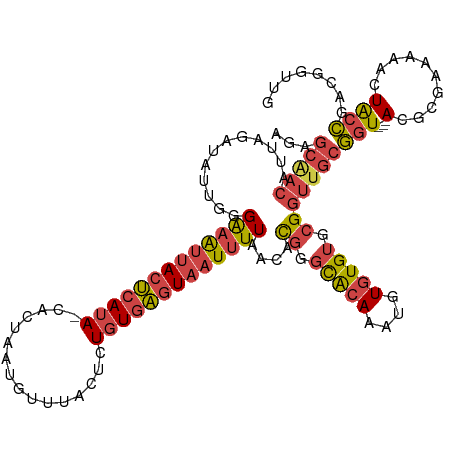
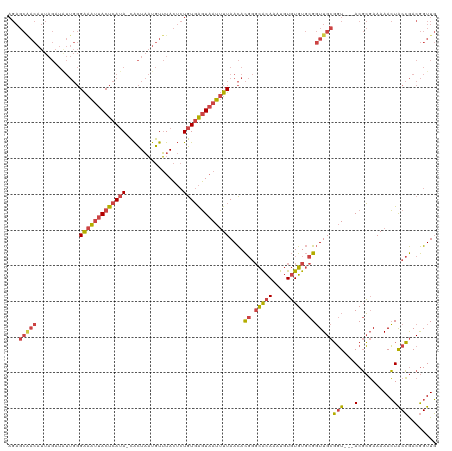
| Location | 15,654,441 – 15,654,557 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.55 |
| Shannon entropy | 0.35745 |
| G+C content | 0.38345 |
| Mean single sequence MFE | -26.27 |
| Consensus MFE | -17.54 |
| Energy contribution | -19.94 |
| Covariance contribution | 2.39 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.972736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
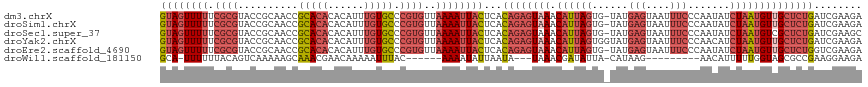
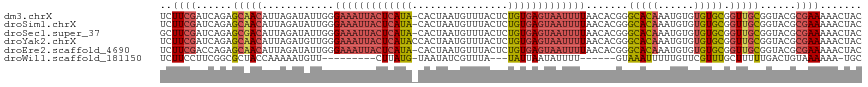
>dm3.chrX 15654441 116 + 22422827 GUAGUUUUUCGCGUACCGCAACCGCACACACAUUUGUGCCCGUGUUAAAAUUACUCACAGAGUAAACAUUAGUG-UAUGAGUAAUUUCCCAAUAUCUAAUGUUGCUCUGAUCGAAGA ((((((((.((((....((....)).((((....))))..))))..))))))))...((((((((.((((((((-(..(((....)))...))).))))))))))))))........ ( -30.10, z-score = -3.23, R) >droSim1.chrX 12136720 116 + 17042790 GUAGUUUUUCGCGUACCGCAACCGCACACACAUUUGUGCCCGUGUUAAAAUUACUCACAGAGUAAACAUUAGUG-UAUGAGUAAUUUCCCAAUAUCUAAUGUUGCUCUGAUCGAAGA ((((((((.((((....((....)).((((....))))..))))..))))))))...((((((((.((((((((-(..(((....)))...))).))))))))))))))........ ( -30.10, z-score = -3.23, R) >droSec1.super_37 238654 116 + 454039 GUAGUUUUUCGCGUACCGCAACCGCACACACAUUUGUGCCCGUGUUAAAAUUACUCACAGAGUAAACAUUAGUG-UAUGAGUAAUUUCCCAAUAUCUAAUGUCGCUCUGAUCGAAGC ((((((((.((((....((....)).((((....))))..))))..))))))))...((((((..(((((((((-(..(((....)))...))).))))))).))))))........ ( -28.20, z-score = -2.59, R) >droYak2.chrX 9859468 117 + 21770863 GUAGUUUUUCGCGUACCGCAACCGCACACACAUUUGUGCCCGUGUUAAAAUUACUCACAGAGUAAACAUUAGUGGUAUGAGUAAUUUCCCAACAUCUAAUGUUGCUCUGAUCGAAGA ((((((((.((((....((....)).((((....))))..))))..))))))))...((((((((.((((((((....(((....)))....)).))))))))))))))........ ( -31.60, z-score = -2.95, R) >droEre2.scaffold_4690 7364870 116 - 18748788 GUAGUUUUUCGCGUACCGCAACCGCACACACAUUUGUGCCCGUGUUAAAAUUACUCACAGAGUAAACAUUAGUG-UAUGAGUAAUUUCCCAAUAUCUAAUGUUGCUCUGGUCGAAGA ((((((((.((((....((....)).((((....))))..))))..))))))))...((((((((.((((((((-(..(((....)))...))).))))))))))))))........ ( -30.30, z-score = -2.73, R) >droWil1.scaffold_181150 2480672 97 + 4952429 GCA-UUUUUUACAGUCAAAAAGCAAACGAACAAAAAUUUAC------AAAAUAUUAAUA---UAAACGAUAUUA-CAUAAG---------AACAUUUUUGGUAGCGCCGAAGGAAGA ((.-(((((.......)))))))..................------.......(((((---(.....))))))-......---------....(((((((.....))))))).... ( -7.30, z-score = 0.69, R) >consensus GUAGUUUUUCGCGUACCGCAACCGCACACACAUUUGUGCCCGUGUUAAAAUUACUCACAGAGUAAACAUUAGUG_UAUGAGUAAUUUCCCAAUAUCUAAUGUUGCUCUGAUCGAAGA ((((((((.((((..........(((((......))))).))))..))))))))...((((((((.((((((......(((....))).......))))))))))))))........ (-17.54 = -19.94 + 2.39)
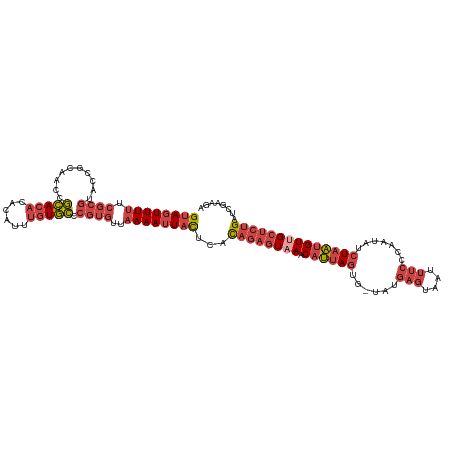
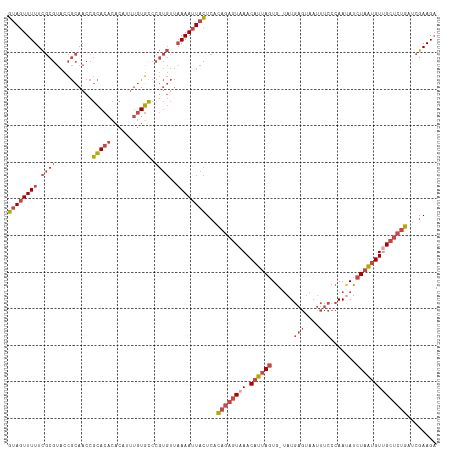
| Location | 15,654,441 – 15,654,557 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.55 |
| Shannon entropy | 0.35745 |
| G+C content | 0.38345 |
| Mean single sequence MFE | -28.12 |
| Consensus MFE | -20.35 |
| Energy contribution | -21.51 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.936944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15654441 116 - 22422827 UCUUCGAUCAGAGCAACAUUAGAUAUUGGGAAAUUACUCAUA-CACUAAUGUUUACUCUGUGAGUAAUUUUAACACGGGCACAAAUGUGUGUGCGGUUGCGGUACGCGAAAAACUAC ..((((..(((((.((((((((....((((......))))..-..))))))))..)))))...(((....((((.((.(((((....))))).))))))...))).))))....... ( -31.60, z-score = -2.60, R) >droSim1.chrX 12136720 116 - 17042790 UCUUCGAUCAGAGCAACAUUAGAUAUUGGGAAAUUACUCAUA-CACUAAUGUUUACUCUGUGAGUAAUUUUAACACGGGCACAAAUGUGUGUGCGGUUGCGGUACGCGAAAAACUAC ..((((..(((((.((((((((....((((......))))..-..))))))))..)))))...(((....((((.((.(((((....))))).))))))...))).))))....... ( -31.60, z-score = -2.60, R) >droSec1.super_37 238654 116 - 454039 GCUUCGAUCAGAGCGACAUUAGAUAUUGGGAAAUUACUCAUA-CACUAAUGUUUACUCUGUGAGUAAUUUUAACACGGGCACAAAUGUGUGUGCGGUUGCGGUACGCGAAAAACUAC ..((((..(((((.((((((((....((((......))))..-..))))))))..)))))...(((....((((.((.(((((....))))).))))))...))).))))....... ( -32.00, z-score = -2.27, R) >droYak2.chrX 9859468 117 - 21770863 UCUUCGAUCAGAGCAACAUUAGAUGUUGGGAAAUUACUCAUACCACUAAUGUUUACUCUGUGAGUAAUUUUAACACGGGCACAAAUGUGUGUGCGGUUGCGGUACGCGAAAAACUAC ..((((......(((((......((((..(((((((((((((................)))))))))))))))))((.(((((....))))).)))))))......))))....... ( -32.59, z-score = -2.13, R) >droEre2.scaffold_4690 7364870 116 + 18748788 UCUUCGACCAGAGCAACAUUAGAUAUUGGGAAAUUACUCAUA-CACUAAUGUUUACUCUGUGAGUAAUUUUAACACGGGCACAAAUGUGUGUGCGGUUGCGGUACGCGAAAAACUAC ..((((.((((((.((((((((....((((......))))..-..))))))))..))))).).(((....((((.((.(((((....))))).))))))...))).))))....... ( -32.00, z-score = -2.47, R) >droWil1.scaffold_181150 2480672 97 - 4952429 UCUUCCUUCGGCGCUACCAAAAAUGUU---------CUUAUG-UAAUAUCGUUUA---UAUUAAUAUUUU------GUAAAUUUUUGUUCGUUUGCUUUUUGACUGUAAAAAA-UGC ........((((.......((((((((---------...(((-(((......)))---))).))))))))------((((((........)))))).....).))).......-... ( -8.90, z-score = 0.59, R) >consensus UCUUCGAUCAGAGCAACAUUAGAUAUUGGGAAAUUACUCAUA_CACUAAUGUUUACUCUGUGAGUAAUUUUAACACGGGCACAAAUGUGUGUGCGGUUGCGGUACGCGAAAAACUAC ..((((......(((((............(((((((((((((................))))))))))))).......(((((......))))).)))))......))))....... (-20.35 = -21.51 + 1.16)
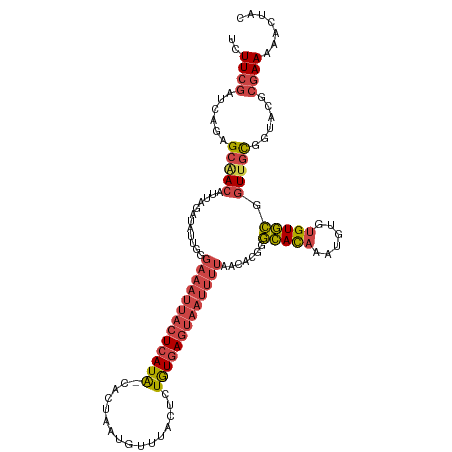
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:47:10 2011