| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,596,123 – 15,596,236 |
| Length | 113 |
| Max. P | 0.659612 |

| Location | 15,596,123 – 15,596,236 |
|---|---|
| Length | 113 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.72 |
| Shannon entropy | 0.68105 |
| G+C content | 0.52478 |
| Mean single sequence MFE | -31.36 |
| Consensus MFE | -14.28 |
| Energy contribution | -14.29 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.61 |
| Mean z-score | -0.79 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.659612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
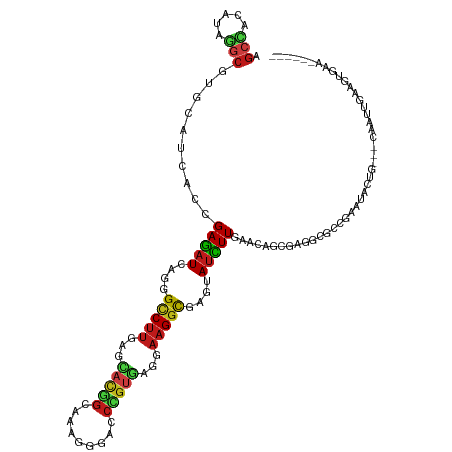
>dm3.chrX 15596123 113 + 22422827 CGCCACGUAGGCGUGCAUCACCGAGAUCAGGGCCUUGAGCACGGCAAAGGGACCCGUGAGGAAGGCGAGUAUCUUGAAGAGCGAUGCGCCGAAUACUG---CAAUUGAAGUCCAUC---- .(((.....)))(((((((.(((((((....(((((...(((((.........)))))...)))))....))))))....).))))))).........---...............---- ( -33.10, z-score = 0.39, R) >droSim1.chrX 12093180 113 + 17042790 CGCCACGUAGGCGUGCAUCACCGAGAUCAGGGCCUUGAGCACGGCAAAGGGACCCGUGAGGAAGGCGAGUAUCUUGAAGAGCGAUGCGCCGAAUACUG---CAAUUGAAGUCCAUC---- .(((.....)))(((((((.(((((((....(((((...(((((.........)))))...)))))....))))))....).))))))).........---...............---- ( -33.10, z-score = 0.39, R) >droSec1.super_37 179662 113 + 454039 CGCCACGUAGGCGUGCAUCACCGAGAUCAGGGCCUUGAGCACGGCAAAGGGACCCGUGAGGAAGGCGAGUAUCUUGAAGAGCGAUGCGCCGAAUACUG---CAAUUGAAGUCCAUC---- .(((.....)))(((((((.(((((((....(((((...(((((.........)))))...)))))....))))))....).))))))).........---...............---- ( -33.10, z-score = 0.39, R) >droYak2.chrX 9801239 113 + 21770863 CGCCACAUAGGCGUGCAUCACCGAGAUCAAUGCCUUAAGCACGGCAAAGGGACCCGUGAGGAAGGCGAGUAUCUUGAAAAGCGAUGCGCCGAAUACUG---CAAUUGAAGGGUACA---- .(((.....)))(((((((.(((((((...((((((...(((((.........)))))...))))))...))))))....).))))))).........---...............---- ( -35.20, z-score = -1.21, R) >droEre2.scaffold_4690 7306593 113 - 18748788 CGCCACAUAGGCGUGCAUCACCGAGAUCAGGGCCUUGAGCACGGCAAAGGGACCCGUGAGGAAGGCGAGUAUCUUGAAGAGCGAAGCGCCGAAUACUG---CAAUUGAAGAGUACA---- .(((.....)))((((.((.((....((((((((((..((...))..)))).))).)))....)).))...((((.((..(((.............))---)..)).)))))))).---- ( -32.22, z-score = -0.21, R) >droAna3.scaffold_12929 612180 103 - 3277472 CGCCACAUAGGCGUGCAUCACCGAGAUCAGAGCCUUCAGCACGGCGAAGGGGCCCGUGAGGAAGGCGAGAAUCUUGAACAGCGAGGCCCCGAAUACUA---CAAGA-------------- ((((.....))))(((.....((((((....((((((..((((((......).)))))..))))))....))))))....)))...............---.....-------------- ( -33.00, z-score = -1.12, R) >dp4.chrXL_group1a 1168760 120 - 9151740 AGCCACAUAGGCGUGCAUCACGGAGAUCAAGGCCUUCAGCACGGCAAAGGGGCCCGUGAGGAAGGCCAGUAUUUUGAAGAGCGAGGCGCCGAAUACUAUCACAGUUCAAGCGAAAGUUCA .(((.....)))(((((((.....)))...(((((((..((((((......).)))))..))))))).))))......((((....(((.((((.........))))..)))...)))). ( -38.70, z-score = -1.73, R) >droPer1.super_15 1841552 120 - 2181545 AGCCACAUAGGCGUGCAUCACGGAGAUCAAGGCCUUCAGCACGGCAAAGGGGCCCGUGAGGAAGGCCAGUAUUUUGAAGAGCGAGGCGCCGAAUACUAUCACAGUUCAAGCGAGAGUUCA .(((.....)))(((((((.....)))...(((((((..((((((......).)))))..))))))).))))......((((....(((.((((.........))))..)))...)))). ( -38.70, z-score = -1.59, R) >droWil1.scaffold_181150 1598996 111 + 4952429 AGCUACAUAAGCAUGCAUCACUGAGAUCAAUGCCUUCAAUACCGCAAACGGUCCACUGAGGAAGGCUAAAAUUUUAAACAACGAUGCUCCAAGUACUG---UAGAAAAAAGGAA------ ..(((((((.....(((((..((((((....((((((...((((....))))........))))))....))))))......)))))......)).))---)))..........------ ( -23.40, z-score = -1.18, R) >droVir3.scaffold_13042 1633298 112 + 5191987 AGCGACAUAGGCAUGCAUAACAGAGAUCAGGGCCUUUAGCACCGCAAAGGGCCCGGUAAGGAAGGCAAGUAUUUUGAACAGCGAAACGCCGAUGACUG---CAAACAAAAUUAAA----- .(((.(((.(((.(((.........(((.((((((((.((...)).))))))))))).......)))....(((((.....))))).))).)))..))---).............----- ( -28.99, z-score = -1.40, R) >droMoj3.scaffold_6328 3515146 112 + 4453435 AGCUACGUAGGCGUGCAUCACAGAGAUCAGAGCCUUCAGUAUAGCGAAGGGGCCCGUGAGAAAGGCCAAAAUCUUAUACAACGAAGCGCCUAAUACUG---GUGAGAAAAAAAUA----- .((((..(((((((........(((((.....(((((........)))))(((((....)...))))...)))))..........)))))))....))---))............----- ( -29.77, z-score = -1.38, R) >droGri2.scaffold_15081 2570593 112 + 4274704 AGCCACAUAGGCAUGCAUUACAGAGAUCAAUGCCUUGAGCACCGCAAAAGGUGCGGUGAGGAAGGCCAAUAUUUUGAACAACGAGGCGCCGAAUACUG---CAAAUAAAGAUGCG----- .(((.....)))..(((((............((((((..(((((((.....))))))).....(..(((....)))..)..))))))((........)---).......))))).----- ( -33.50, z-score = -2.27, R) >anoGam1.chr2R 39900376 92 + 62725911 GGCAACGAACCCGUGAAUCACCGAGAUGCCCGUCUUGACGACGGCCACCGGGAACGAGACGAUGGCGAGCAGUUUAAACGUGGAGAAGCCCA---------------------------- (((..(...(((((.....((.(...(((((((((((....(((...)))....)))))))..))))..).))....))).)).)..)))..---------------------------- ( -25.40, z-score = 0.00, R) >triCas2.ChLG6 3733138 108 + 13544221 AGCUACAACGAGGUGAAGGAGGGAAAUCCCCGACUUAACGAUGGCAACAGGCGCUGAUAAGUAGAUGAUGAUUUUAAACAGCGAAAUCCCAACUGGAA---UAAUUCAAUU--------- .(((........((.(((..(((.....)))..))).))..((....)))))((((.....((((.......))))..))))....(((.....))).---..........--------- ( -20.90, z-score = -0.15, R) >consensus AGCCACAUAGGCGUGCAUCACCGAGAUCAGGGCCUUGAGCACGGCAAAGGGACCCGUGAGGAAGGCGAGUAUCUUGAACAGCGAGGCGCCGAAUACUG___CAAUUGAAGUGAA______ .(((.....)))..........(((((....(((((...(((((.........)))))...)))))....)))))............................................. (-14.28 = -14.29 + 0.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:47:03 2011