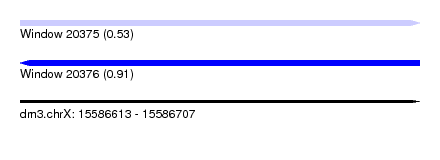
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,586,613 – 15,586,707 |
| Length | 94 |
| Max. P | 0.911403 |

| Location | 15,586,613 – 15,586,707 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 72.15 |
| Shannon entropy | 0.51754 |
| G+C content | 0.42028 |
| Mean single sequence MFE | -21.39 |
| Consensus MFE | -10.95 |
| Energy contribution | -11.06 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.526824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
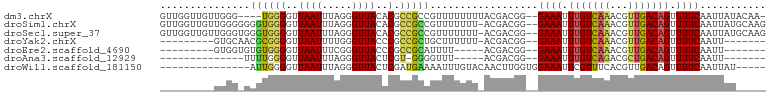
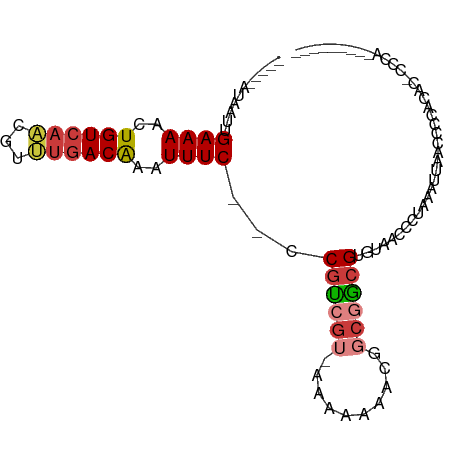
>dm3.chrX 15586613 94 + 22422827 GUUGGUUGUUGGG----UGGGGUUAAUUUAGGGUUACACGCCGCCGUUUUUUUUACGACGG--GAAAUUUGUCAAACGUUGACAGUUUUCAAUUAUACAA- ....(((((..((----(((.((((((.....)))).)).))))).........)))))..--((((.((((((.....)))))).))))..........- ( -21.60, z-score = -0.55, R) >droSim1.chrX 12079531 98 + 17042790 GUUGGUUGUUGGGGGGGUGGGGUUAAUUUAGGGUUACACGCCGCCGUUUUUUU-ACGACGG--GAAAUUUGUCAAACGUUGACAGUUUUCAAUUAUGCAAG .((((((((.((((.(((((.((((((.....)))).)).))))).))))...-)))))..--((((.((((((.....)))))).)))).......))). ( -27.40, z-score = -2.07, R) >droSec1.super_37 166627 98 + 454039 GUUGGUUGUUGGGUGGGUGGGGUUAAUUUAGGGUUACACGCCGCCGUUUUUUU-ACGACGG--GAAAUUUGUCAAACGUUGACAGUUUUCAAUUAUGCAAG ((((((((..((.(((((((.((((((.....)))).)).)))))........-.(((((.--((......))...))))).)).))..)))))).))... ( -24.30, z-score = -0.82, R) >droYak2.chrX 9788090 82 + 21770863 ---------GUGCAACGCGGGGUUAAUUUUGGGUUACCCGCCGCUGCUUUUUU-ACGACGG--GAAAUUUGUCAAACGUUGACAGUUUUCAAUU------- ---------..(((..((((((.((((.....)))).)).)))))))......-.......--((((.((((((.....)))))).))))....------- ( -21.10, z-score = -0.50, R) >droEre2.scaffold_4690 7293527 78 - 18748788 ---------GUGGUGUGUGGGGUUAAUUUCGGGUUACCCGCCGCAUUUU-----ACGACGG--GAAAUUUGUCAAACGUUGACAGUUUUCAAUU------- ---------((((.((((((((.((((.....)))).)).)))))).))-----)).....--((((.((((((.....)))))).))))....------- ( -23.00, z-score = -1.43, R) >droAna3.scaffold_12929 596526 72 - 3277472 --------------UUUUGGGGUUAAUUUAGGGUUACUCGU-GGGGUUU-----ACGACGG--GAAAUUUGUCAGACGCUGACAGUUUUCAAUU------- --------------..(((..(..............(((((-.(.....-----.).))))--)....(((((((...))))))).)..)))..------- ( -16.40, z-score = -1.66, R) >droWil1.scaffold_181150 1581826 81 + 4952429 ---------------AUUGGGGUUAAUUUAGGGUUACUCGAUGAAAAUUUGUACAACUUGGUGGAAAUUCGUUUCACGUUGACAGUUUUCAAUUAU----- ---------------....((((.(((.....)))))))..((((((((.((.((((...((((((.....)))))))))))))))))))).....----- ( -15.90, z-score = -1.12, R) >consensus _________UGGG_GGGUGGGGUUAAUUUAGGGUUACACGCCGCCGUUUUUUU_ACGACGG__GAAAUUUGUCAAACGUUGACAGUUUUCAAUUAU_____ ...............((((((..((((.....))))..).)))))..................((((.(((((((...))))))).))))........... (-10.95 = -11.06 + 0.11)

| Location | 15,586,613 – 15,586,707 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 72.15 |
| Shannon entropy | 0.51754 |
| G+C content | 0.42028 |
| Mean single sequence MFE | -16.09 |
| Consensus MFE | -7.53 |
| Energy contribution | -8.14 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.911403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
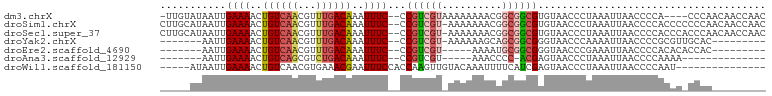
>dm3.chrX 15586613 94 - 22422827 -UUGUAUAAUUGAAAACUGUCAACGUUUGACAAAUUUC--CCGUCGUAAAAAAAACGGCGGCGUGUAACCCUAAAUUAACCCCA----CCCAACAACCAAC -((((......((((..(((((.....)))))..))))--(((((((.......))))))).......................----....))))..... ( -16.60, z-score = -1.36, R) >droSim1.chrX 12079531 98 - 17042790 CUUGCAUAAUUGAAAACUGUCAACGUUUGACAAAUUUC--CCGUCGU-AAAAAAACGGCGGCGUGUAACCCUAAAUUAACCCCACCCCCCCAACAACCAAC .((((((....((((..(((((.....)))))..))))--(((((((-......))))))).))))))................................. ( -19.90, z-score = -2.91, R) >droSec1.super_37 166627 98 - 454039 CUUGCAUAAUUGAAAACUGUCAACGUUUGACAAAUUUC--CCGUCGU-AAAAAAACGGCGGCGUGUAACCCUAAAUUAACCCCACCCACCCAACAACCAAC .((((((....((((..(((((.....)))))..))))--(((((((-......))))))).))))))................................. ( -19.90, z-score = -2.65, R) >droYak2.chrX 9788090 82 - 21770863 -------AAUUGAAAACUGUCAACGUUUGACAAAUUUC--CCGUCGU-AAAAAAGCAGCGGCGGGUAACCCAAAAUUAACCCCGCGUUGCAC--------- -------....((((..(((((.....)))))..))))--.......-......((((((.((((...............))))))))))..--------- ( -21.46, z-score = -1.96, R) >droEre2.scaffold_4690 7293527 78 + 18748788 -------AAUUGAAAACUGUCAACGUUUGACAAAUUUC--CCGUCGU-----AAAAUGCGGCGGGUAACCCGAAAUUAACCCCACACACCAC--------- -------..(((.....(((((.....))))).....(--(((((((-----.....)))))))).....)))...................--------- ( -17.50, z-score = -1.43, R) >droAna3.scaffold_12929 596526 72 + 3277472 -------AAUUGAAAACUGUCAGCGUCUGACAAAUUUC--CCGUCGU-----AAACCCC-ACGAGUAACCCUAAAUUAACCCCAAAA-------------- -------....((((..((((((...))))))..))))--...((((-----.......-)))).......................-------------- ( -9.50, z-score = -2.51, R) >droWil1.scaffold_181150 1581826 81 - 4952429 -----AUAAUUGAAAACUGUCAACGUGAAACGAAUUUCCACCAAGUUGUACAAAUUUUCAUCGAGUAACCCUAAAUUAACCCCAAU--------------- -----.....((((((.((((((((((...........)))...)))).)))..))))))..........................--------------- ( -7.80, z-score = -0.52, R) >consensus _____AUAAUUGAAAACUGUCAACGUUUGACAAAUUUC__CCGUCGU_AAAAAAACGGCGGCGUGUAACCCUAAAUUAACCCCACAC_CCCA_________ ...........((((..((((((...))))))..))))...((((((..........))))))...................................... ( -7.53 = -8.14 + 0.62)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:47:00 2011