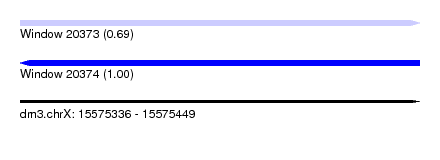
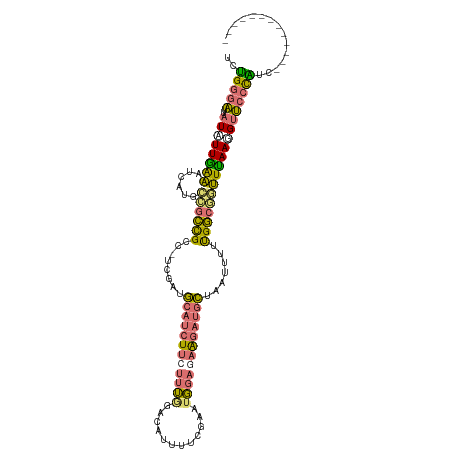
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,575,336 – 15,575,449 |
| Length | 113 |
| Max. P | 0.999417 |

| Location | 15,575,336 – 15,575,449 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 59.45 |
| Shannon entropy | 0.64902 |
| G+C content | 0.40404 |
| Mean single sequence MFE | -24.35 |
| Consensus MFE | -9.76 |
| Energy contribution | -11.20 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.692157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15575336 113 + 22422827 AAAAAAUGGCAUGGGAAGGAACAUUGAAUGGCCAAAAAUUAGCAUCUUUUACAUUCGAAAAUGUCAAAAGAAGAUGCAUCAA-AGCGGCGGCAUGAUUUCAAUAUAAUCCCCGA ............((((......((((((..(((........((((((((((((((....)))))....))))))))).((..-...)).))).....))))))....))))... ( -23.60, z-score = -0.47, R) >droSim1.chrX 12069617 101 + 17042790 -------------GAUGGGAACCUUAAACCGCCAAAAAUUAGCAUCUUCUCCAUUCGAAAAUGUCCAAAGAAGAUGCAUCGGCGGCGGCGGCAUGAUUUCAAUAUAAUCCCAGA -------------..(((((........(((((........(((((((((.((((....)))).....)))))))))...(....))))))..((....))......))))).. ( -27.80, z-score = -1.75, R) >droSec1.super_37 153540 100 + 454039 -------------GAUGGGAACCUUAAACCGCCAGAAAUUAGCAUCUUCUCCAUUCGAAAAUGUCCAAAGAAGAUGCAUCAA-GGCGGCGGCAUGAUUUCAAUAUAAUCCCAGA -------------..(((((........(((((........(((((((((.((((....)))).....))))))))).....-...)))))..((....))......))))).. ( -27.69, z-score = -2.34, R) >droWil1.scaffold_181150 3375737 103 - 4952429 -------AAGUAAAACAGGUGCGUUAAUGAGUCGAAAAUGGACUUUCAAUCAAUUGACAUAUAUUUCAC--AGGUUUAAGGUGGGCAAACCUUAGAAACUAACAGCAAAGCG-- -------............(((((((((..((.((((......)))).))..))))))...........--...((((((((......))))))))........))).....-- ( -18.30, z-score = -0.50, R) >consensus _____________GAUAGGAACCUUAAACCGCCAAAAAUUAGCAUCUUCUCCAUUCGAAAAUGUCCAAAGAAGAUGCAUCAA_GGCGGCGGCAUGAUUUCAAUAUAAUCCCAGA ...............(((((........(((((........(((((((((..................))))))))).........)))))..((....))......))))).. ( -9.76 = -11.20 + 1.44)

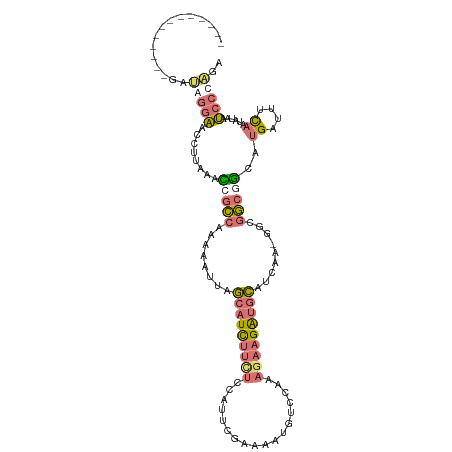
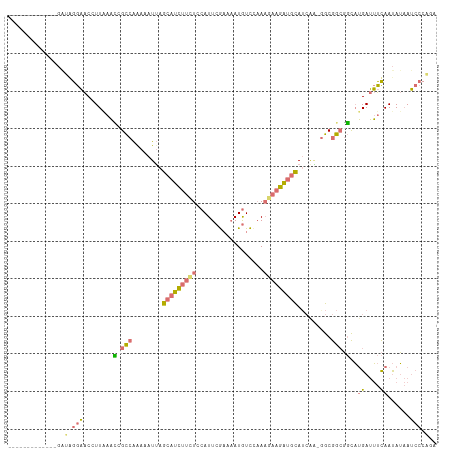
| Location | 15,575,336 – 15,575,449 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 59.45 |
| Shannon entropy | 0.64902 |
| G+C content | 0.40404 |
| Mean single sequence MFE | -28.61 |
| Consensus MFE | -19.08 |
| Energy contribution | -18.96 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.58 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.87 |
| SVM RNA-class probability | 0.999417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15575336 113 - 22422827 UCGGGGAUUAUAUUGAAAUCAUGCCGCCGCU-UUGAUGCAUCUUCUUUUGACAUUUUCGAAUGUAAAAGAUGCUAAUUUUUGGCCAUUCAAUGUUCCUUCCCAUGCCAUUUUUU ..(((((..(((((((....(((..((((..-.....(((((((......(((((....)))))..))))))).......))))))))))))))...)))))............ ( -26.94, z-score = -1.66, R) >droSim1.chrX 12069617 101 - 17042790 UCUGGGAUUAUAUUGAAAUCAUGCCGCCGCCGCCGAUGCAUCUUCUUUGGACAUUUUCGAAUGGAGAAGAUGCUAAUUUUUGGCGGUUUAAGGUUCCCAUC------------- ..(((((......((....)).(((...((((((((.(((((((((..(..(......)..)..)))))))))......))))))))....))))))))..------------- ( -36.10, z-score = -3.22, R) >droSec1.super_37 153540 100 - 454039 UCUGGGAUUAUAUUGAAAUCAUGCCGCCGCC-UUGAUGCAUCUUCUUUGGACAUUUUCGAAUGGAGAAGAUGCUAAUUUCUGGCGGUUUAAGGUUCCCAUC------------- ..(((((......((....)).(((((((((-..((.(((((((((..(..(......)..)..))))))))).....)).))))))....))))))))..------------- ( -33.50, z-score = -2.86, R) >droWil1.scaffold_181150 3375737 103 + 4952429 --CGCUUUGCUGUUAGUUUCUAAGGUUUGCCCACCUUAAACCU--GUGAAAUAUAUGUCAAUUGAUUGAAAGUCCAUUUUCGACUCAUUAACGCACCUGUUUUACUU------- --....................(((((((.......)))))))--((((((((..(((....((((((((((....))))))).))).....)))..))))))))..------- ( -17.90, z-score = -1.22, R) >consensus UCUGGGAUUAUAUUGAAAUCAUGCCGCCGCC_UCGAUGCAUCUUCUUUGGACAUUUUCGAAUGGAGAAGAUGCUAAUUUUUGGCGGUUUAAGGUUCCCAUC_____________ ..(((((..((((((((......((((((........((((((((((((............)))))))))))).......)))))))))))))))))))............... (-19.08 = -18.96 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:46:59 2011