| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,571,475 – 15,571,566 |
| Length | 91 |
| Max. P | 0.756532 |

| Location | 15,571,475 – 15,571,566 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 64.38 |
| Shannon entropy | 0.76163 |
| G+C content | 0.65198 |
| Mean single sequence MFE | -34.47 |
| Consensus MFE | -12.58 |
| Energy contribution | -13.35 |
| Covariance contribution | 0.77 |
| Combinations/Pair | 1.38 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.553867 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
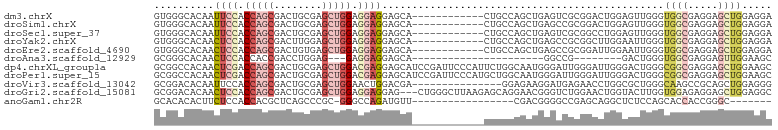
>dm3.chrX 15571475 91 + 22422827 GUGGGCACAAUUCCACCAGCGACUGCGAGCUGGAGGAGGAGCA------------CUGCCAGCUGAGUCGCGGACUGGAGUUGGGUGGCGAGGAGCUGGAGGA ....((.((((((((((.((((((.(.((((((((........------------)).)))))))))))))))..)))))))).))(((.....)))...... ( -36.50, z-score = -1.11, R) >droSim1.chrX 12065845 91 + 17042790 GUGGGCACAAUUCCACCAGCGACUGCGAGCUGGAGGAGGAGCA------------CUGCCAGCUGAGCCGCGGACUGGAGUUGGGUGGCGAGGAGCUGGAGGA ....((....((((.(((((........))))).))))..)).------------((.((((((..(((((.(((....))).).))))....)))))).)). ( -36.30, z-score = -0.83, R) >droSec1.super_37 149746 91 + 454039 GUGGGCACAAUUCCACCAGCGACUGCGAGCUGGAGGAGGAGCA------------CUGCCAGCUGAGUCGCGGCCUGGAGUUGGGUGGCGAGGAGCUGGAGGA ....((....((((.(((((........))))).))))..)).------------((.((((((...((((.((((......)))).))))..)))))).)). ( -40.20, z-score = -1.52, R) >droYak2.chrX 9766598 91 + 21770863 GUGGGCACAACUCCACCAGCGACUUGGAGCUGGAGGAGGAGCA------------CUGCCAGCUGAGCCGCGGCUUGGAAUUGGGUGGCGAGGAGCUGGAGGA ....((....((((.(((((........))))).))))..)).------------((.((((((....(((.((((......)))).)))...)))))).)). ( -37.50, z-score = -1.21, R) >droEre2.scaffold_4690 7278535 91 - 18748788 GUGGGCACAACUCCACCAGCGACUGUGAGCUGGAGGAGGAGCA------------CUGCCAGCUGAGCCGCGGAUUGGAAUUGGGUGGCGAGGAGCUGGAGGA ....((....((((.(((((........))))).))))..)).------------((.((((((..(((((.............)))))....)))))).)). ( -36.52, z-score = -1.56, R) >droAna3.scaffold_12929 578278 70 - 3277472 GCGGGCACAACUCCACCACCGACCUGGAG---GAGGAGGAGCA----------------------GGCCG--------GACUGGGUGGCGAGGAGUUGGAAGC ((...(.(((((((.(((((..(((....---.))).....((----------------------(....--------..))))))))...))))))))..)) ( -25.90, z-score = -0.79, R) >dp4.chrXL_group1a 1146289 103 - 9151740 GCGGCCACAACUCGACCAGCGACUGCGAGCUGGACGAGGAGCAUCCGAUUCCCAUUCUGGCAAUGGGAUUGGGAUUGGGACUGGGCGGCGAGGAGCUGGAAGC ((.(((.((.((((.(((((........))))).)))).....(((((((((.(((((......))))).)))))))))..))))).)).............. ( -41.50, z-score = -1.91, R) >droPer1.super_15 1818709 103 - 2181545 GCGGCCACAACUCGACCAGCGACUGCGAGCUGGACGAGGAGCAUCCGAUUCCCAUGCUGGCAAUGGGAUUGGGAUUGGGACUGGGCGGCGAGGAGCUGGAAGC ((.(((.((.((((.(((((........))))).))))....((((.(.((((((.......)))))).).))))......))))).)).............. ( -40.20, z-score = -1.45, R) >droVir3.scaffold_13042 1598167 88 + 5191987 GCGGACACAAUUCCACCAGCGACUGCGAGCUGGAACUGGACGA---------------GGAGAAGGAUGAGAACCUGGCGCUGGGCAAGCCGCAGCUGGAGGG ............((.(((((...((((.(((....(..(.((.---------------.....(((.......)))..)))..)...)))))))))))).)). ( -27.20, z-score = -0.04, R) >droGri2.scaffold_15081 2549770 100 + 4274704 GCGGACACAACUCCACCAGCGACUGCGAGCUGGAGGAGGAG---CUGGGCUUAAGAGCAGGAACGGGUCUGGAACUGGUACUUGGUGGAGAGGAGCUGGAGGC .(((((....((((.(((((........))))).))))...---(((..(((......)))..))))))))...(..((.(((......)))..))..).... ( -33.20, z-score = -0.49, R) >anoGam1.chr2R 36355598 78 - 62725911 GCACACACUUCUCCACCACGCUCAGCCCGC-GGGCCAGAUGUU-----------------CGACGGGGCCGAGCAGGCUCUCCAGCACCACCGGGC------- ...............((..((((.((((.(-((((.....)))-----------------))...)))).))))..(((....)))......))..------- ( -24.20, z-score = 0.42, R) >consensus GCGGGCACAACUCCACCAGCGACUGCGAGCUGGAGGAGGAGCA____________CUGCCAGAUGGGCCGCGGACUGGAAUUGGGUGGCGAGGAGCUGGAGGA ..........((((.(((((........))))).))))...............................................((((.....))))..... (-12.58 = -13.35 + 0.77)

| Location | 15,571,475 – 15,571,566 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 64.38 |
| Shannon entropy | 0.76163 |
| G+C content | 0.65198 |
| Mean single sequence MFE | -29.83 |
| Consensus MFE | -9.24 |
| Energy contribution | -9.83 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.756532 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 15571475 91 - 22422827 UCCUCCAGCUCCUCGCCACCCAACUCCAGUCCGCGACUCAGCUGGCAG------------UGCUCCUCCUCCAGCUCGCAGUCGCUGGUGGAAUUGUGCCCAC .......((.(....(((((..((....))..(((((((((((((.((------------........)))))))).).)))))).)))))....).)).... ( -28.80, z-score = -1.69, R) >droSim1.chrX 12065845 91 - 17042790 UCCUCCAGCUCCUCGCCACCCAACUCCAGUCCGCGGCUCAGCUGGCAG------------UGCUCCUCCUCCAGCUCGCAGUCGCUGGUGGAAUUGUGCCCAC ....((((((....(((.(...((....))..).)))..))))))..(------------..(...(((.(((((........))))).)))...)..).... ( -28.80, z-score = -1.17, R) >droSec1.super_37 149746 91 - 454039 UCCUCCAGCUCCUCGCCACCCAACUCCAGGCCGCGACUCAGCUGGCAG------------UGCUCCUCCUCCAGCUCGCAGUCGCUGGUGGAAUUGUGCCCAC ....((((((..((((..((........))..))))...))))))..(------------..(...(((.(((((........))))).)))...)..).... ( -32.00, z-score = -1.89, R) >droYak2.chrX 9766598 91 - 21770863 UCCUCCAGCUCCUCGCCACCCAAUUCCAAGCCGCGGCUCAGCUGGCAG------------UGCUCCUCCUCCAGCUCCAAGUCGCUGGUGGAGUUGUGCCCAC ....((((((....(((.(.............).)))..))))))..(------------..(..((((.(((((........))))).))))..)..).... ( -31.62, z-score = -2.25, R) >droEre2.scaffold_4690 7278535 91 + 18748788 UCCUCCAGCUCCUCGCCACCCAAUUCCAAUCCGCGGCUCAGCUGGCAG------------UGCUCCUCCUCCAGCUCACAGUCGCUGGUGGAGUUGUGCCCAC ....((((((....(((.(.............).)))..))))))..(------------..(..((((.(((((........))))).))))..)..).... ( -31.62, z-score = -2.73, R) >droAna3.scaffold_12929 578278 70 + 3277472 GCUUCCAACUCCUCGCCACCCAGUC--------CGGCC----------------------UGCUCCUCCUC---CUCCAGGUCGGUGGUGGAGUUGUGCCCGC ((...(((((((..((((((.....--------.((((----------------------((.........---...))))))))))))))))))).)).... ( -26.40, z-score = -2.54, R) >dp4.chrXL_group1a 1146289 103 + 9151740 GCUUCCAGCUCCUCGCCGCCCAGUCCCAAUCCCAAUCCCAUUGCCAGAAUGGGAAUCGGAUGCUCCUCGUCCAGCUCGCAGUCGCUGGUCGAGUUGUGGCCGC ((.....)).....((.(((........((((...(((((((.....)))))))...))))((..((((.(((((........))))).))))..))))).)) ( -36.80, z-score = -2.30, R) >droPer1.super_15 1818709 103 + 2181545 GCUUCCAGCUCCUCGCCGCCCAGUCCCAAUCCCAAUCCCAUUGCCAGCAUGGGAAUCGGAUGCUCCUCGUCCAGCUCGCAGUCGCUGGUCGAGUUGUGGCCGC ((.....)).....((.(((........((((...((((((.(....)))))))...))))((..((((.(((((........))))).))))..))))).)) ( -35.90, z-score = -1.89, R) >droVir3.scaffold_13042 1598167 88 - 5191987 CCCUCCAGCUGCGGCUUGCCCAGCGCCAGGUUCUCAUCCUUCUCC---------------UCGUCCAGUUCCAGCUCGCAGUCGCUGGUGGAAUUGUGUCCGC .(((...((((.((....))))))...)))...............---------------..(.(((((((((.((.((....)).)))))))))).).)... ( -24.70, z-score = -0.23, R) >droGri2.scaffold_15081 2549770 100 - 4274704 GCCUCCAGCUCCUCUCCACCAAGUACCAGUUCCAGACCCGUUCCUGCUCUUAAGCCCAG---CUCCUCCUCCAGCUCGCAGUCGCUGGUGGAGUUGUGUCCGC .......((.(..((((((((.((..(.....(((........))).......((..((---((........)))).)).)..))))))))))..).)).... ( -22.10, z-score = -0.24, R) >anoGam1.chr2R 36355598 78 + 62725911 -------GCCCGGUGGUGCUGGAGAGCCUGCUCGGCCCCGUCG-----------------AACAUCUGGCCC-GCGGGCUGAGCGUGGUGGAGAAGUGUGUGC -------..((((.....))))...((((((((((((((((((-----------------......))))..-).)))))))))).))).............. ( -29.40, z-score = 0.68, R) >consensus UCCUCCAGCUCCUCGCCACCCAAUUCCAGUCCCCGACCCAGCUGGCAG____________UGCUCCUCCUCCAGCUCGCAGUCGCUGGUGGAGUUGUGCCCGC ..................................................................(((.(((((........))))).)))........... ( -9.24 = -9.83 + 0.59)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:46:57 2011