
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,566,558 – 15,566,661 |
| Length | 103 |
| Max. P | 0.865380 |

| Location | 15,566,558 – 15,566,655 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 73.23 |
| Shannon entropy | 0.47371 |
| G+C content | 0.56406 |
| Mean single sequence MFE | -29.19 |
| Consensus MFE | -20.06 |
| Energy contribution | -20.58 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.865380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
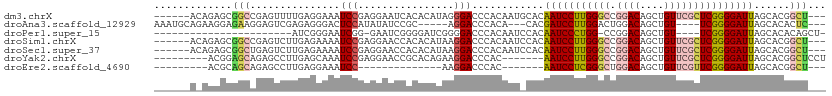
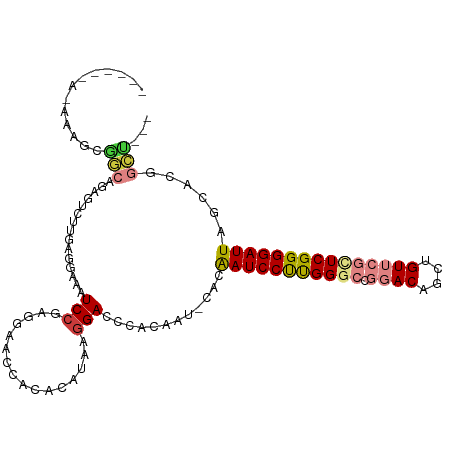
>dm3.chrX 15566558 97 - 22422827 CGGCCGAGUUUUGAGGAAAUCCGAGGAAUCACACAUAGGGACCCACAAUGCACAAUCCUUGGGCCGGACAGCUGUUCGCUCGGGGAUUAGCACGGCU .(((((.(((((..((....))..)))))...................(((..(((((((((((.((((....))))))))))))))).)))))))) ( -33.10, z-score = -1.45, R) >droAna3.scaffold_12929 572806 83 + 3277472 --AAGGAGUCGAGAGGGACUCCAUAUAUCCGCA-----GGACCC---ACACACGAUCCUUGGACUGGACAGCUGU----UCGGGGAUUAGCACACUC --..((((((......))))))........((.-----(....)---......(((((((((((.........))----))))))))).))...... ( -25.60, z-score = -1.12, R) >droSim1.chrX 12061263 97 - 17042790 CGGCCGAGUCUUGAGAAAAUCCGAGGAACCACACAUAAGGACCCACAAUCCACAAUCCUUGGGCCGGACAGCUGUUCGCUCGGGGAUUAGCACGGCU .(((((..(((((.(.....))))))............(((.......)))..(((((((((((.((((....)))))))))))))))....))))) ( -31.20, z-score = -1.19, R) >droSec1.super_37 144853 97 - 454039 CGGCUGAGUCUUGAGAAAAUCCGAGGAACCACACAUAAGGACCCACAAUCCACAAUCCUUGGGCCGGACAGCUGUUCGCUCGGGGAUUAGCACGGCU (((((((..((((((....((((((((...........(((.......)))....)))))))).(((((....)))))))))))..))))).))... ( -29.46, z-score = -0.73, R) >droEre2.scaffold_4690 7273533 73 + 18748788 ---CAGAGCCUUGAGGAAAUCCAAGG---------------------ACCCACAAUCCUCGGGCUGGACAGCUGUUCGUUCGGGGAUUAGCACGGCU ---...((((.((.((....))....---------------------......(((((((((((.((((....)))))))))))))))..)).)))) ( -26.60, z-score = -1.49, R) >consensus CGGCCGAGUCUUGAGGAAAUCCGAGGAACCACACAUA_GGACCCACAAUCCACAAUCCUUGGGCCGGACAGCUGUUCGCUCGGGGAUUAGCACGGCU ..((((.((((((........))))............................(((((((((((.((((....))))))))))))))).)).)))). (-20.06 = -20.58 + 0.52)



| Location | 15,566,558 – 15,566,661 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 66.99 |
| Shannon entropy | 0.63218 |
| G+C content | 0.57139 |
| Mean single sequence MFE | -30.49 |
| Consensus MFE | -15.82 |
| Energy contribution | -16.92 |
| Covariance contribution | 1.10 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.83 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.756976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15566558 103 - 22422827 ------ACAGAGCGGCCGAGUUUUGAGGAAAUCCGAGGAAUCACACAUAGGGACCCACAAUGCACAAUCCUUGGGCCGGACAGCUGUUCGCUCGGGGAUUAGCACGGCU--- ------.......(((((.(((((..((....))..)))))...................(((..(((((((((((.((((....))))))))))))))).))))))))--- ( -33.10, z-score = -0.99, R) >droAna3.scaffold_12929 572806 97 + 3277472 AAAUGCAGAAGGAGAAGGAGUCGAGAGGGACUCCAUAUAUCCGC-----AGGACCCACA---CACGAUCCUUGGACUGGACAGCUGU----UCGGGGAUUAGCACACUC--- ...(((....(((...((((((......)))))).....)))..-----..........---...(((((((((((.........))----))))))))).))).....--- ( -29.60, z-score = -1.36, R) >droPer1.super_15 1813168 82 + 2181545 -----------------------AUCGGGAAUCGG-GAAUCGGGGAUCGGGGACCCACAAUCCACAAUCCCUGG-CCGGACAGCUGU----UCGGGGAUUAGCACACAGCU- -----------------------......((((..-...((((((((.(.(((.......))).).))))))))-((((((....))----)))).))))(((.....)))- ( -25.40, z-score = 0.31, R) >droSim1.chrX 12061263 103 - 17042790 ------ACAGAGCGGCCGAGUCUUGAGAAAAUCCGAGGAACCACACAUAAGGACCCACAAUCCACAAUCCUUGGGCCGGACAGCUGUUCGCUCGGGGAUUAGCACGGCU--- ------.......(((((..(((((.(.....))))))............(((.......)))..(((((((((((.((((....)))))))))))))))....)))))--- ( -31.20, z-score = -0.69, R) >droSec1.super_37 144853 103 - 454039 ------ACAGAGCGGCUGAGUCUUGAGAAAAUCCGAGGAACCACACAUAAGGACCCACAAUCCACAAUCCUUGGGCCGGACAGCUGUUCGCUCGGGGAUUAGCACGGCU--- ------....(((.(((((..((((((....((((((((...........(((.......)))....)))))))).(((((....)))))))))))..)))))...)))--- ( -30.06, z-score = -0.37, R) >droYak2.chrX 9761588 96 - 21770863 ---------ACGGAGCAGAGCCUUGAGCAAAUCCGAGGAACCGCACAGAAGGACCCAC-------AAUCCUUGGGCCGGACAGCUGUUCGCUCGGGGAUUAGCACGGCUCCU ---------..(((((....(((((........)))))....((......(....)..-------(((((((((((.((((....))))))))))))))).))...))))). ( -36.00, z-score = -1.50, R) >droEre2.scaffold_4690 7273533 79 + 18748788 ---------ACGCAGCAGAGCCUUGAGGAAAUCC--------------AAGGACCCAC-------AAUCCUCGGGCUGGACAGCUGUUCGUUCGGGGAUUAGCACGGCU--- ---------..((.((....(((((........)--------------))))......-------(((((((((((.((((....))))))))))))))).))...)).--- ( -28.10, z-score = -1.17, R) >consensus ______A_AAAGCGGCAGAGUCUUGAGGAAAUCCGAGGAACCACACAUAAGGACCCACAAU_CACAAUCCUUGGGCCGGACAGCUGUUCGCUCGGGGAUUAGCACGGCU___ .............(((.(.((..........(((................)))............(((((((((((.((((....))))))))))))))).)).).)))... (-15.82 = -16.92 + 1.10)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:46:55 2011