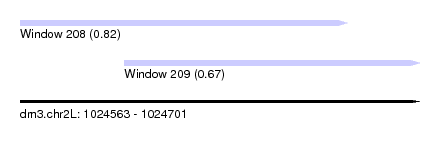
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,024,563 – 1,024,701 |
| Length | 138 |
| Max. P | 0.818358 |

| Location | 1,024,563 – 1,024,676 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 71.71 |
| Shannon entropy | 0.51209 |
| G+C content | 0.39455 |
| Mean single sequence MFE | -19.98 |
| Consensus MFE | -13.10 |
| Energy contribution | -13.35 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.818358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
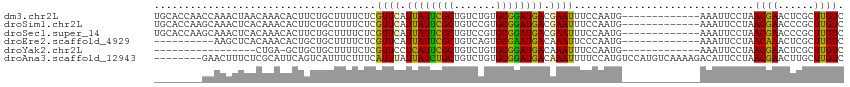
>dm3.chr2L 1024563 113 + 23011544 ACAAGUCUUUUCUGA----UUCAGAAACUUAUCUUUAAUUUGCACCAACCAAACUAACAAACACUUCUGCUUUUCUCGUUCAUUAUUCGCUGUCUGUGCGGAUGACGAAUUUCCAAU ..((((...(((((.----..)))))))))...............................................((((.((((((((.......)))))))).))))....... ( -14.90, z-score = -0.25, R) >droSim1.chr2L 994977 113 + 22036055 ACAAGUCUUUUCUGA----UUCAGAAACUUAUCUGUAAGUUGCACCAAGCAAACUCACAAACACUUCUGCUUUUCUCGUUCAUUAUUCGCUGUCCGUGCGGAUGACGAAUUUCCAAU ..((((..((((((.----..))))))......(((.((((((.....)).)))).))).........)))).....((((.((((((((.......)))))))).))))....... ( -20.90, z-score = -0.84, R) >droSec1.super_14 977462 113 + 2068291 ACAAGCCUUUUCUGA----UUCAGAAACUUAUCUGUAAGUUGCACCAAGCAAACUCACAAACACUUCUGCUUUUCUCGUUCAUUAUUCGCUGUCCGUGCGGAUGACGAAUUUCCAAU ..((((..((((((.----..))))))......(((.((((((.....)).)))).))).........)))).....((((.((((((((.......)))))))).))))....... ( -22.80, z-score = -1.49, R) >droEre2.scaffold_4929 1067302 97 + 26641161 ACAAGUUCUCUCUGA----UUCAGAAAGUUGUCUA----------------AGCUCACAAACACUGCUGCUUUUCUCGUUCAUUAUUCGCUGUCAGUGCGAAUGACAAAUUCCCAAU .......((.((((.----..)))).))(((((.(----------------(((...((.....))..))))............((((((.......)))))))))))......... ( -15.00, z-score = 0.35, R) >droYak2.chr2L 1000379 89 + 22324452 GCGAGUCCUUUUUGC----UUCAGAAACUUGUCU-------------------CUGA-----GCUGCUGCUUUUCUCGUUCCUCAUUCGCUGUCUGUGCGGAUGACAAAUUUCCAAU (((((........((----(.((((........)-------------------))))-----))..........)))))...((((((((.......))))))))............ ( -22.27, z-score = -1.69, R) >droAna3.scaffold_12943 3537407 107 + 5039921 GCAGGUUUUCAUUGCAAAUUUAAGAAACUUUUUUGU----------GAGAACUUUCUCGCAUUCAGUCAUUUCUUUCAUUUAUUAUCUGCUGUCUGUGCGGAUGACAAAUUUUCCAU ((((.......))))((((..((((((((....(((----------((((....)))))))...)))...)))))..)))).((((((((.......))))))))............ ( -24.00, z-score = -2.08, R) >consensus ACAAGUCUUUUCUGA____UUCAGAAACUUAUCUGU__________AAGCAAACUCACAAACACUGCUGCUUUUCUCGUUCAUUAUUCGCUGUCUGUGCGGAUGACAAAUUUCCAAU ........(((((((.....)))))))..................................................((((.((((((((.......)))))))).))))....... (-13.10 = -13.35 + 0.25)

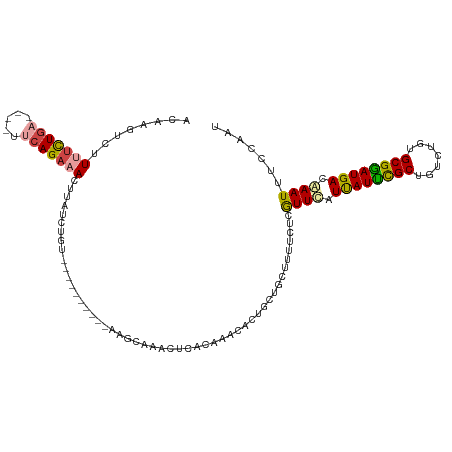
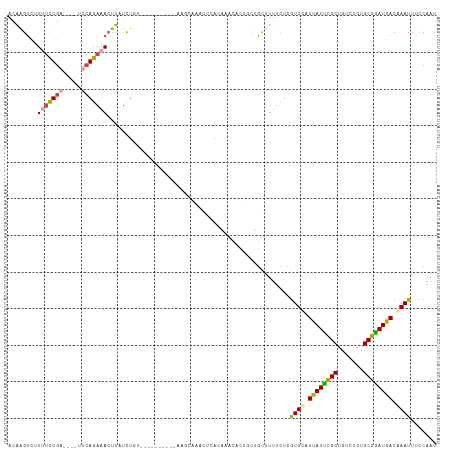
| Location | 1,024,599 – 1,024,701 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 75.14 |
| Shannon entropy | 0.43350 |
| G+C content | 0.43067 |
| Mean single sequence MFE | -18.13 |
| Consensus MFE | -11.38 |
| Energy contribution | -10.72 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.666464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1024599 102 + 23011544 UGCACCAACCAAACUAACAAACACUUCUGCUUUUCUCGUUCAUUAUUCGCUGUCUGUGCGGAUGACGAAUUUCCAAUG-------------AAAUUCCUAACGAACUCGCUUGUC .....................................((((.((((((((.......)))))))).(((((((....)-------------)))))).....))))......... ( -17.70, z-score = -1.45, R) >droSim1.chr2L 995013 102 + 22036055 UGCACCAAGCAAACUCACAAACACUUCUGCUUUUCUCGUUCAUUAUUCGCUGUCCGUGCGGAUGACGAAUUUCCAAUG-------------AAAUUCCUAACGAACCCGCUUGUC .....(((((.......((........))........((((.((((((((.......)))))))).(((((((....)-------------)))))).....))))..))))).. ( -22.00, z-score = -2.24, R) >droSec1.super_14 977498 102 + 2068291 UGCACCAAGCAAACUCACAAACACUUCUGCUUUUCUCGUUCAUUAUUCGCUGUCCGUGCGGAUGACGAAUUUCCAAUG-------------AAAUUCCUAACGAACCCGCUUGUC .....(((((.......((........))........((((.((((((((.......)))))))).(((((((....)-------------)))))).....))))..))))).. ( -22.00, z-score = -2.24, R) >droEre2.scaffold_4929 1067332 92 + 26641161 ----------AAGCUCACAAACACUGCUGCUUUUCUCGUUCAUUAUUCGCUGUCAGUGCGAAUGACAAAUUCCCAAUG-------------AAAUUCCUAACAAACUCGCUUGUC ----------((((............................((((((((.......))))))))....(((.....)-------------))...............))))... ( -11.50, z-score = 0.33, R) >droYak2.chr2L 1000409 84 + 22324452 -----------------CUGA-GCUGCUGCUUUUCUCGUUCCUCAUUCGCUGUCUGUGCGGAUGACAAAUUUCCAAUG-------------AAAUUCCUAACGAACUCGCUUGUC -----------------..((-((....))))...(((((..((((((((.......))))))))..((((((....)-------------)))))...)))))........... ( -18.20, z-score = -1.67, R) >droAna3.scaffold_12943 3537445 107 + 5039921 --------GAACUUUCUCGCAUUCAGUCAUUUCUUUCAUUUAUUAUCUGCUGUCUGUGCGGAUGACAAAUUUUCCAUGUCCAUGUCAAAAGACAUUCCUAACGAACUUGCUUGUC --------..........((((((.............((((.((((((((.......)))))))).))))...........(((((....))))).......)))..)))..... ( -17.40, z-score = -0.40, R) >consensus ________GCAAACUCACAAACACUGCUGCUUUUCUCGUUCAUUAUUCGCUGUCUGUGCGGAUGACAAAUUUCCAAUG_____________AAAUUCCUAACGAACUCGCUUGUC .....................................((((.((((((((.......)))))))).))))..............................((((......)))). (-11.38 = -10.72 + -0.66)
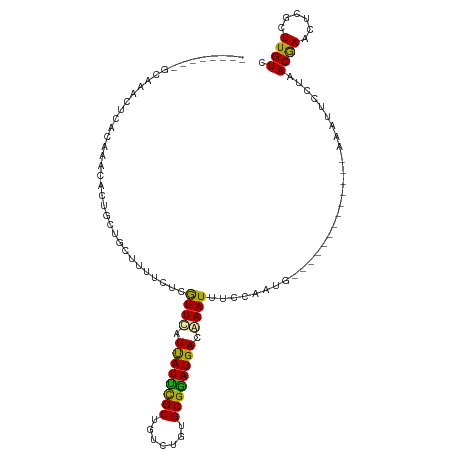
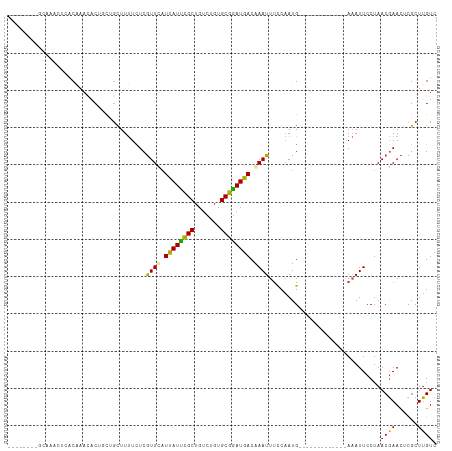
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:07:33 2011