| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,516,568 – 15,516,647 |
| Length | 79 |
| Max. P | 0.938914 |

| Location | 15,516,568 – 15,516,647 |
|---|---|
| Length | 79 |
| Sequences | 11 |
| Columns | 79 |
| Reading direction | reverse |
| Mean pairwise identity | 79.48 |
| Shannon entropy | 0.45258 |
| G+C content | 0.53629 |
| Mean single sequence MFE | -26.12 |
| Consensus MFE | -15.60 |
| Energy contribution | -15.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.938914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
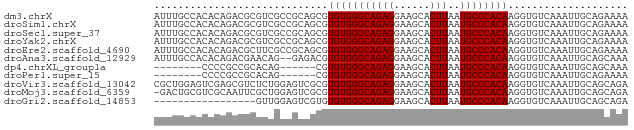
>dm3.chrX 15516568 79 - 22422827 AUUUGCCACACAGACGCGUCGCCGCAGCGUGUGGGCAGAGGAAGCACUUAAUGCCCACAAGGUGUCAAAUUGCAGAAAA .(((((......(((((..(((....)))(((((((((((......)))..))))))))..))))).....)))))... ( -28.00, z-score = -1.96, R) >droSim1.chrX 11998812 79 - 17042790 AUUUGCCACACAGACGCGUCGCCGCAGCGUGUGGGCAGAGGAAGCACUUAAUGCCCACAAGGUGUCAAAUUGCAGAAAA .(((((......(((((..(((....)))(((((((((((......)))..))))))))..))))).....)))))... ( -28.00, z-score = -1.96, R) >droSec1.super_37 94531 79 - 454039 AUUUGCCACACAGACGCGUCGCCGCAGCGUGUGGGCAGAGGAAGCACUUAAUGCCCACAAGGUGUCAAAUUGCAGAAAA .(((((......(((((..(((....)))(((((((((((......)))..))))))))..))))).....)))))... ( -28.00, z-score = -1.96, R) >droYak2.chrX 9705773 79 - 21770863 AUUUGCCACACAGACGCGUCGCCGCAGCGUGUGGGCAGAGGAAGCACUUAAUGCCCACAAGGUGUCAAAUUGCAGAAAA .(((((......(((((..(((....)))(((((((((((......)))..))))))))..))))).....)))))... ( -28.00, z-score = -1.96, R) >droEre2.scaffold_4690 7224420 79 + 18748788 AUUUGCCACACAGACGCUUCGCCGCAGCGUGUGGGCAGAGGAAGCACUUAAUGCCCACAAGGUGUCAAAUUGCAGAAAA .(((((......((((((((((....)))(((((((((((......)))..))))))))))))))).....)))))... ( -28.90, z-score = -2.43, R) >droAna3.scaffold_12929 503762 77 + 3277472 AUUUGCCACACAGACGAACAG--GAGACGUGUGGGCAGAGGAAGCACUUAAUGCCCACAAGGUGUCAAAUUGCAGCAAA .(((((..((..((((....(--....).(((((((((((......)))..))))))))...))))....))..))))) ( -23.70, z-score = -2.12, R) >dp4.chrXL_group1a 8537697 65 - 9151740 --------CCCCGCCGCACAG------CGUGUGGGCAGAGGAAGCACUUAAUGCCCACAAGGUGUCAAAUUGCAGCAAA --------....((.(((((.------(.(((((((((((......)))..)))))))).).)))......)).))... ( -22.80, z-score = -1.50, R) >droPer1.super_15 933283 65 - 2181545 --------CCCCGCCGCACAG------CGUGUGGGCAGAGGAAGCACUUAAUGCCCACAAGGUGUCAAAUUGCAGAAAA --------...((((((...)------).(((((((((((......)))..)))))))).))))............... ( -20.70, z-score = -1.22, R) >droVir3.scaffold_13042 1175048 79 + 5191987 CGCUGGAGUCGAGCGUCUCUGGAGUCGCGUGUGGGCAGAGGAAGCACUUAAUGCCCACAAGGUGUCAAAUUGCAGCAGA .((((.(((...(((.((....)).))).(((((((((((......)))..)))))))).........))).))))... ( -29.60, z-score = -1.57, R) >droMoj3.scaffold_6359 2376401 78 - 4525533 -GACUGCGUCGCAAUUCGCUGGAGUCGCGUGUGGGCAGAGGAAGCACUUAAUGCCCACAAGGUGUCAAAUUGCAGCAGA -..((((...((((((...(((..((...(((((((((((......)))..)))))))).))..))))))))).)))). ( -29.90, z-score = -1.89, R) >droGri2.scaffold_14853 6474852 62 - 10151454 -----------------GUUGGAGUCGUGUGUGGGCAGAGGAAGCACUUAAUGCCCACAAGGUGUCAAAUUGCAGCAGA -----------------((((.(((((..(((((((((((......)))..))))))))...))....))).))))... ( -19.70, z-score = -1.52, R) >consensus AUUUGCCACACAGACGCGUCGCCGCAGCGUGUGGGCAGAGGAAGCACUUAAUGCCCACAAGGUGUCAAAUUGCAGAAAA .............................(((((((((((......)))..)))))))).................... (-15.60 = -15.60 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:46:52 2011