
| Sequence ID | dm3.chrX |
|---|---|
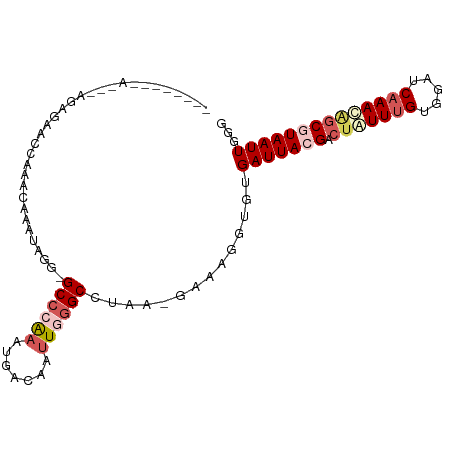
| Location | 15,511,220 – 15,511,313 |
| Length | 93 |
| Max. P | 0.998572 |

| Location | 15,511,220 – 15,511,313 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 73.10 |
| Shannon entropy | 0.44437 |
| G+C content | 0.41095 |
| Mean single sequence MFE | -24.47 |
| Consensus MFE | -12.74 |
| Energy contribution | -14.42 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.977731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15511220 93 + 22422827 --------AGCCAGAGAACCAAACAAAUAGA-GCCCAAAUGACAAUUGGGCCUAAAGAAAGGUGUGAUUACGACUAUUUGUUGAGCAAACAGCGUAAUUGGG --------..((.....(((.......(((.-((((((.......)))))))))......)))..(((((((.((.((((.....)))).))))))))))). ( -24.32, z-score = -2.84, R) >droEre2.scaffold_4690 7219358 93 - 18748788 --------AAAUAAAUAAAGGGAGAAUUAAGUGCCCGCAUGCCAAUUAGGCGCAG-GAAAGGUGUGAUUACGAAUAUUUGUAGAUCAAAAGGCAUAAUUGGG --------.........................((((.(((((......((((..-.....))))((((((((....)))).))))....)))))...)))) ( -17.10, z-score = -0.33, R) >droYak2.chrX 9700662 101 + 21770863 UCCCAGAAAACUAAACAAAUAAAGGGAGAAAUGCGCGAAUGACAAUUAGGCCUAA-GAAAGGUGUGAUUACGACGAUUUGUGGAUCUAAUGGCAUAAUUGGG ((((...................))))...((.(((((((...(((((.((((..-...)))).)))))......))))))).))(((((......))))). ( -18.91, z-score = -0.77, R) >droSec1.super_37 89464 88 + 454039 ------------AGAGGGCCAAACAAAUAGG-GCCCAAAUGACAAUUGGGCCUAA-GAAAAUUGUGAUUACGACUGUUUGUGGAUCAAACAGCGUAAUUGGU ------------.....(((.........((-((((((.......))))))))..-.........(((((((.(((((((.....))))))))))))))))) ( -31.50, z-score = -4.42, R) >droSim1.chrX 11993711 88 + 17042790 ------------AGAGGGCCAAACAAAUAGG-GCCCAAAUGACAAUUGGGCCUAA-GAAAAUUGUGAUUACGACUGUUUGCGGAUCAAACAGCGUAAUUGGU ------------.....(((.........((-((((((.......))))))))..-.........(((((((.(((((((.....))))))))))))))))) ( -30.50, z-score = -4.03, R) >consensus ________A___AGAGAACCAAACAAAUAGG_GCCCAAAUGACAAUUGGGCCUAA_GAAAGGUGUGAUUACGACUAUUUGUGGAUCAAACAGCGUAAUUGGG ................................((((((.......))))))..............(((((((.(((((((.....))))))))))))))... (-12.74 = -14.42 + 1.68)

| Location | 15,511,220 – 15,511,313 |
|---|---|
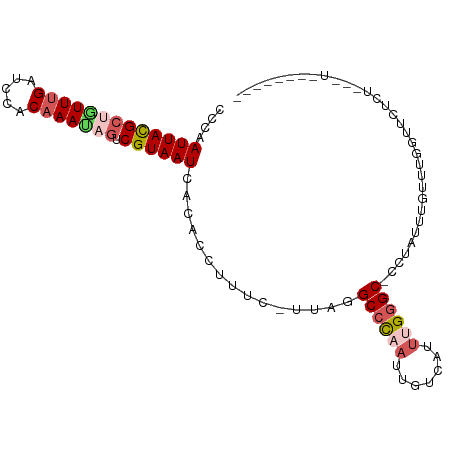
| Length | 93 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 73.10 |
| Shannon entropy | 0.44437 |
| G+C content | 0.41095 |
| Mean single sequence MFE | -22.10 |
| Consensus MFE | -13.44 |
| Energy contribution | -14.48 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.41 |
| SVM RNA-class probability | 0.998572 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
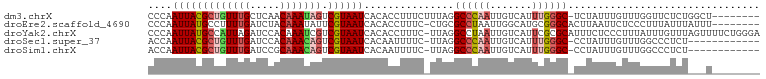
>dm3.chrX 15511220 93 - 22422827 CCCAAUUACGCUGUUUGCUCAACAAAUAGUCGUAAUCACACCUUUCUUUAGGCCCAAUUGUCAUUUGGGC-UCUAUUUGUUUGGUUCUCUGGCU-------- .((((((((((((((((.....))))))).))))))...(((...(..(((((((((.......))))))-.)))...)...)))....)))..-------- ( -27.50, z-score = -4.50, R) >droEre2.scaffold_4690 7219358 93 + 18748788 CCCAAUUAUGCCUUUUGAUCUACAAAUAUUCGUAAUCACACCUUUC-CUGCGCCUAAUUGGCAUGCGGGCACUUAAUUCUCCCUUUAUUUAUUU-------- ........(((((..((((.(((........)))))))........-..(((((.....)))..))))))).......................-------- ( -14.20, z-score = -1.29, R) >droYak2.chrX 9700662 101 - 21770863 CCCAAUUAUGCCAUUAGAUCCACAAAUCGUCGUAAUCACACCUUUC-UUAGGCCUAAUUGUCAUUCGCGCAUUUCUCCCUUUAUUUGUUUAGUUUUCUGGGA ..((((((.(((....(((......)))...((....)).......-...))).))))))...............((((...................)))) ( -12.61, z-score = 0.11, R) >droSec1.super_37 89464 88 - 454039 ACCAAUUACGCUGUUUGAUCCACAAACAGUCGUAAUCACAAUUUUC-UUAGGCCCAAUUGUCAUUUGGGC-CCUAUUUGUUUGGCCCUCU------------ .((((((((((((((((.....))))))).)))))...........-...(((((((.......))))))-)........))))......------------ ( -28.00, z-score = -5.02, R) >droSim1.chrX 11993711 88 - 17042790 ACCAAUUACGCUGUUUGAUCCGCAAACAGUCGUAAUCACAAUUUUC-UUAGGCCCAAUUGUCAUUUGGGC-CCUAUUUGUUUGGCCCUCU------------ .((((((((((((((((.....))))))).)))))...........-...(((((((.......))))))-)........))))......------------ ( -28.20, z-score = -4.79, R) >consensus CCCAAUUACGCUGUUUGAUCCACAAAUAGUCGUAAUCACACCUUUC_UUAGGCCCAAUUGUCAUUUGGGC_CCUAUUUGUUUGGUUCUCU___U________ ....(((((((((((((.....))))))).))))))...............((((((.......))))))................................ (-13.44 = -14.48 + 1.04)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:46:51 2011