
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,507,334 – 15,507,427 |
| Length | 93 |
| Max. P | 0.500000 |

| Location | 15,507,334 – 15,507,427 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 60.28 |
| Shannon entropy | 0.73330 |
| G+C content | 0.43949 |
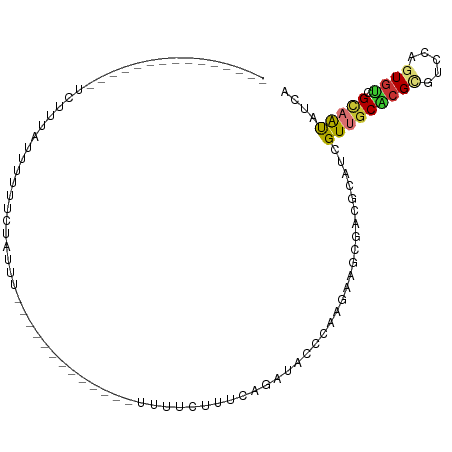
| Mean single sequence MFE | -17.30 |
| Consensus MFE | -6.06 |
| Energy contribution | -6.17 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
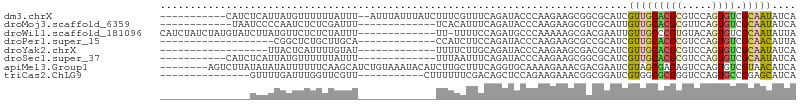
>dm3.chrX 15507334 93 - 22422827 -----------CAUCUCAUUAUGUUUUUUAUUU--AUUUAUUUAUCUUUCGUUUCAGAUACCCAAGAAGCGGCGCAUCGUUGCACGCGUCCAGUGUCGCAAUAUCA -----------......................--.............(((((((..........)))))))......(((((((((.....)))).))))).... ( -14.80, z-score = -0.81, R) >droMoj3.scaffold_6359 2364046 81 - 4525533 ------------UAAUCCCCAAUCUCUCGAUUU-------------UCACAUUUCAGAUACCCAAGAAGCGUCGCAUUGUUGCACGCGUUCAGUGUCGCAAUAUCA ------------........((((....)))).-------------..........(((((....(((((((.(((....))))))).))).)))))......... ( -14.60, z-score = -1.74, R) >droWil1.scaffold_181096 8232522 92 + 12416693 CAUCUAUCUAUGUAUCUUAUGUUCUCUCUAUUU-------------UU-UUUUCCAGAUGCCCAAAAAGCGACGAAUUGUUGCCCGUGUACAGUGUCGCAAUAUUA ...........((((((................-------------..-......)))))).......(((((..(((((.((....))))))))))))....... ( -12.76, z-score = -0.62, R) >droPer1.super_15 923859 74 - 2181545 -------------------CGGCUCUGCUUGCA-------------CCAUCUUCCAGAUACCCAAGAAGCGCCGCAUCGUUGCACGCGUCCAGUGUCGCAACAUUA -------------------((((.((.((((..-------------..(((.....)))...)))).)).))))....(((((((((.....)))).))))).... ( -23.40, z-score = -2.39, R) >droYak2.chrX 9696643 75 - 21770863 ------------------UUACUCAUUUUGUAU-------------UUUUCUUGCAGAUACCCAAGAAGCGACGCAUCGUUGCACGCGUCCAGUGUCGCAAUAUCA ------------------...............-------------..((((((........))))))(((((((..(((.....)))....)))))))....... ( -17.30, z-score = -1.72, R) >droSec1.super_37 85929 82 - 454039 -----------CAUCUCAUUAUGUUUUUUAUUU-------------UUUAAUUUCAGAUACCCAAGAAGCGGCGCAUCGUUGCACGCGUCCAGUGUCGCAAUAUCA -----------......................-------------..........(((((....((.(((..(((....))).))).))..)))))......... ( -13.40, z-score = -0.58, R) >apiMel3.Group1 11078796 98 - 25854376 --------AGUCUUAUAUAUAUUUUUUCAAGCAUCUGUAAAUACAUCUUGCUUUCAGGUGCAAAAGAAACGACGAAUCGUAGCGAGAGUCCAGUGUCGUAACAUCA --------......................(((((((.................))))))).......((((((.......((....))....))))))....... ( -16.73, z-score = 0.01, R) >triCas2.ChLG9 4226491 80 + 15222296 ---------------GUUUUGAUUUGGUUCGUU-----------CUUUUUUCGACAGCUCCAGAAGAAACGGCGGAUCGUGGCGCGGGUCCAGUGCCGGAGCAUCA ---------------(((((.....(((((((.-----------.(((((((..........)))))))..)))))))..(((((.......)))))))))).... ( -25.40, z-score = -0.54, R) >consensus _______________UCUUUAUUUUUUCUAUUU_____________UUUUCUUUCAGAUACCCAAGAAGCGACGCAUCGUUGCACGCGUCCAGUGUCGCAAUAUCA ..............................................................................(((((((((.....)))).))))).... ( -6.06 = -6.17 + 0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:46:50 2011