| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,492,808 – 15,492,873 |
| Length | 65 |
| Max. P | 0.995201 |

| Location | 15,492,808 – 15,492,873 |
|---|---|
| Length | 65 |
| Sequences | 8 |
| Columns | 78 |
| Reading direction | reverse |
| Mean pairwise identity | 59.02 |
| Shannon entropy | 0.77738 |
| G+C content | 0.51055 |
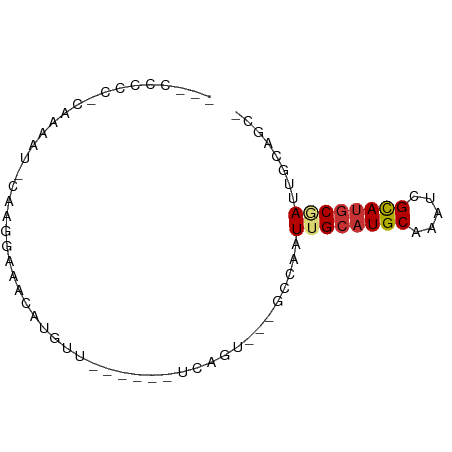
| Mean single sequence MFE | -18.46 |
| Consensus MFE | -8.14 |
| Energy contribution | -8.31 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.995201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
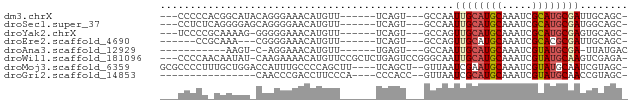
>dm3.chrX 15492808 65 - 22422827 ---CCCCCACGGCAUACAGGGAAACAUGUU------UCAGU---GCCAAUUGCAUGCAAAUCGCAUGCGAUUGCAGC- ---.......((((((((.(....).))).------...))---)))((((((((((.....)))))))))).....- ( -23.80, z-score = -2.71, R) >droSec1.super_37 71649 65 - 454039 ---CCUCUCAGGGGAGCAGGGGAACAUGUU------UCAGU---GCCAAUUGCAUGCAAAUCGCAUGCGAUGGCAGC- ---((.....))((((((.(....).))))------))..(---((((.((((((((.....)))))))))))))..- ( -27.00, z-score = -3.03, R) >droYak2.chrX 9682192 64 - 21770863 ---UCCCCGCAAAAG-GGGGGAAACAUGUU------UCAGU---GCCAGUUGCAUGCAAAUCGCAUGCGAGUGCAGC- ---(((((......)-))))((((....))------))..(---((.(.((((((((.....)))))))).))))..- ( -25.70, z-score = -2.92, R) >droEre2.scaffold_4690 7201125 59 + 18748788 ------CCGCAAA---CGGGGAAACAUGUU------UCAGU---GCCAGUUGCAUGCAAAUCGCACGCGAUUGCAGC- ------..(((((---((.(....).))))------)...(---((.((((((.(((.....))).)))))))))))- ( -17.70, z-score = -0.59, R) >droAna3.scaffold_12929 480893 55 + 3277472 -----------AAGU-C-AGGAAACAUGUU------UGAGU---GCCAAUUGCAUGCAAAUCGUAUGCGA-UUAUGAC -----------..((-(-((....).....------.....---...((((((((((.....))))))))-)).)))) ( -15.80, z-score = -2.38, R) >droWil1.scaffold_181096 8214417 73 + 12416693 ---CCCCAACAAUAU-CAAGAAAACAUGUUCCGCUCUGAGUCCGGGCAAUUGCAUGCAAAUCGUAUGCAAGUCGAGA- ---...........(-(...............((((.......))))..((((((((.....))))))))...))..- ( -14.50, z-score = -0.43, R) >droMoj3.scaffold_6359 2344512 71 - 4525533 GCGCCCCUUUGCUGGACCAUUUGCCCCAGCUU----UCAGCU--GUUAAUCGAAUGCAAAUCGUAUGCAAUCGUAGC- ((((......(((((..(....)..)))))..----...)).--......(((.((((.......)))).)))..))- ( -15.20, z-score = -0.38, R) >droGri2.scaffold_14853 6446601 55 - 10151454 ----------------CAACCCGACCUUCCCA----CCCACC--GUUAAUCGCAUGCAAAUCGUAUGCAACCGUAGC- ----------------................----......--.......((((((.....)))))).........- ( -8.00, z-score = -1.82, R) >consensus ___CCCCC_CAAAAU_CAAGGAAACAUGUU______UCAGU___GCCAAUUGCAUGCAAAUCGCAUGCGAUUGCAGC_ .................................................((((((((.....))))))))........ ( -8.14 = -8.31 + 0.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:46:49 2011