| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,442,957 – 15,443,090 |
| Length | 133 |
| Max. P | 0.998344 |

| Location | 15,442,957 – 15,443,090 |
|---|---|
| Length | 133 |
| Sequences | 5 |
| Columns | 154 |
| Reading direction | forward |
| Mean pairwise identity | 60.57 |
| Shannon entropy | 0.64567 |
| G+C content | 0.43940 |
| Mean single sequence MFE | -35.25 |
| Consensus MFE | -13.44 |
| Energy contribution | -15.40 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.657599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
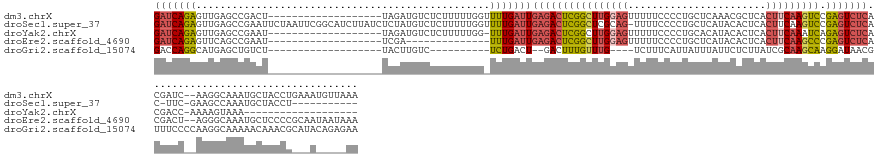
>dm3.chrX 15442957 133 + 22422827 UUUAACAUUUCAGGUAGCAUUUGCCUU--GAUCGUGAGACUCGGACUUGAAGUGAGCGUUUGAGCAGGGGAAAAACUCCAAGCCGAGUCUCAAUCAAAACCAAAAAGAGACAUCUA-------------------AGUCGGCUCAACUCUGAUC .........(((((((((.((((..((--(((..((((((((((.((((......((......)).(((......))))))))))))))))))))))...))))....(((.....-------------------.))).)))..)).)))).. ( -40.80, z-score = -2.33, R) >droSec1.super_37 18192 140 + 454039 -----------AGGUAGCAUUUGGCUUC-GAA-GUGAGACUCGGACUUGAAGUGAGUGUAUGAGCAGGGGAAAA-CUGCGAGCCGAGUCUCAAUCAAAACCAAAAAGAGACAUAGAGAUAAGAUGCCGAAUUAGAAUUCGGCUCAACUCUGAUC -----------..((..(.(((((.((.-((.-.((((((((((((((.....))))......((((.......-))))...)))))))))).)).)).)))))..)..)).(((((....((.(((((((....)))))))))..)))))... ( -45.70, z-score = -3.19, R) >droYak2.chrX 9627322 114 + 21770863 -------------------UUUACUUUU-GGUCGUGAGACUCUGAUUUGAAGUGAGUGUAUGUGCAGGGGAAAAACUCCAAGCCGAGUCUCAAUCAAA-CCAAAAAGAGACAUCUA-------------------AUUCGGCUCAACUCUGAUC -------------------...(((((.-((((....)))).......)))))((((.........((((.....)))).(((((((((((.......-.......))))).....-------------------..))))))..))))..... ( -27.45, z-score = -0.37, R) >droEre2.scaffold_4690 7156207 119 - 18748788 UUUAUUAUUGCGGGGAGCAUUUGCCCU--AGUCGUGAGACUCGGGCUUGAAGUGAGUGUAUGAGCAGGGGAAAAACUCCAAGCCGAGUCUCAAUCAAA--------------UCGA-------------------AUUCGGCUGAACUCUGAUC ..........(((((.((((((((((.--((((....)))).)))).......)))))).......((((.....)))).((((((((.((.......--------------..))-------------------))))))))...)))))... ( -38.51, z-score = -1.65, R) >droGri2.scaffold_15074 6835339 119 + 7742996 UUCUCUGUAUGCGUUUGUUUUUGCCUUGGGGAAACGUUAUCCUUGCUUGCGAUAAGAGAAUAAAUAAUGAAAGA----CAAACAAAGUC--AGUCAGA----------GACAAGUA-------------------AGACAGCUCAUGCCUGGUC .((((((..(((((((((((((.((....))...((((((.....(((.......))).....)))))))))))----)))))...).)--)..))))----------))......-------------------.(((.((....))...))) ( -23.80, z-score = 0.76, R) >consensus UU_____U___AGGUAGCAUUUGCCUU__GGUCGUGAGACUCGGACUUGAAGUGAGUGUAUGAGCAGGGGAAAAACUCCAAGCCGAGUCUCAAUCAAA_CCAAAAAGAGACAUCUA___________________AUUCGGCUCAACUCUGAUC ..................................((((((((((.((((.(((.....................))).)))))))))))))).............................................((((.......)))).. (-13.44 = -15.40 + 1.96)
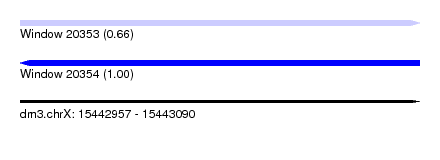
| Location | 15,442,957 – 15,443,090 |
|---|---|
| Length | 133 |
| Sequences | 5 |
| Columns | 154 |
| Reading direction | reverse |
| Mean pairwise identity | 60.57 |
| Shannon entropy | 0.64567 |
| G+C content | 0.43940 |
| Mean single sequence MFE | -36.92 |
| Consensus MFE | -15.30 |
| Energy contribution | -16.14 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.33 |
| SVM RNA-class probability | 0.998344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
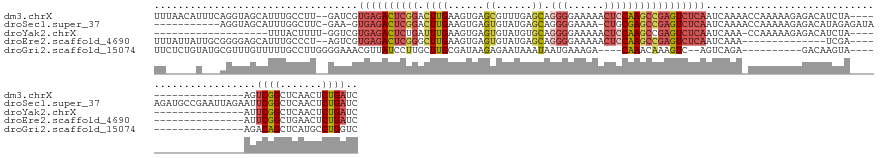
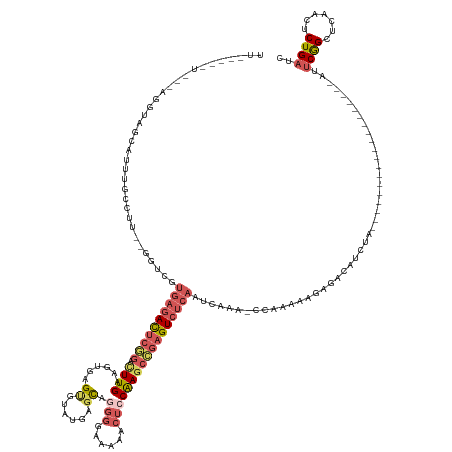
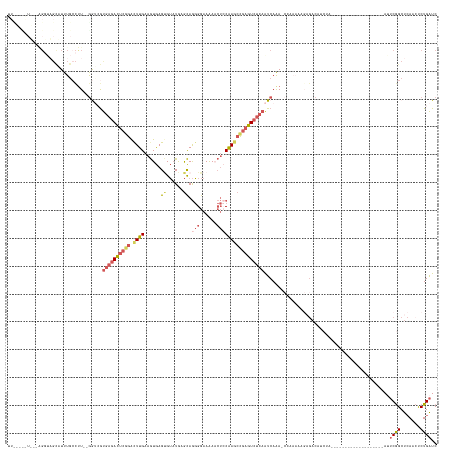
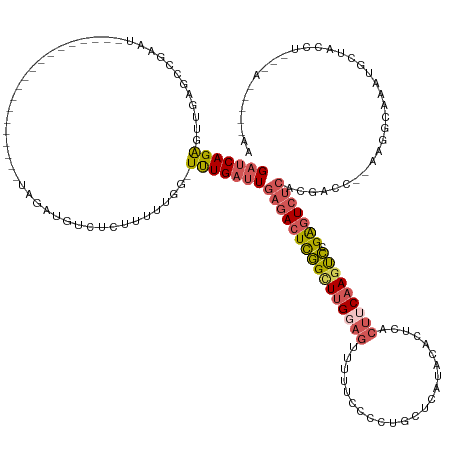
>dm3.chrX 15442957 133 - 22422827 GAUCAGAGUUGAGCCGACU-------------------UAGAUGUCUCUUUUUGGUUUUGAUUGAGACUCGGCUUGGAGUUUUUCCCCUGCUCAAACGCUCACUUCAAGUCCGAGUCUCACGAUC--AAGGCAAAUGCUACCUGAAAUGUUAAA ..((((.((..(((.(((.-------------------.....)))........((((((((((((((((((((((((((.........((......))..)))))))).))))))))))..)))--)))))....)))))))))......... ( -44.50, z-score = -3.26, R) >droSec1.super_37 18192 140 - 454039 GAUCAGAGUUGAGCCGAAUUCUAAUUCGGCAUCUUAUCUCUAUGUCUCUUUUUGGUUUUGAUUGAGACUCGGCUCGCAG-UUUUCCCCUGCUCAUACACUCACUUCAAGUCCGAGUCUCAC-UUC-GAAGCCAAAUGCUACCU----------- (((.((((..(((((((((....))))))).))....))))..)))....(((((((((((.((((((((((...((((-.......))))......(((.......))))))))))))).-.))-)))))))))........----------- ( -49.00, z-score = -5.87, R) >droYak2.chrX 9627322 114 - 21770863 GAUCAGAGUUGAGCCGAAU-------------------UAGAUGUCUCUUUUUGG-UUUGAUUGAGACUCGGCUUGGAGUUUUUCCCCUGCACAUACACUCACUUCAAAUCAGAGUCUCACGACC-AAAAGUAAA------------------- (.((..(((..(((((((.-------------------.(((....))).)))))-))..)))((((((((..(((((((.....................)))))))..).)))))))..)).)-.........------------------- ( -30.00, z-score = -1.28, R) >droEre2.scaffold_4690 7156207 119 + 18748788 GAUCAGAGUUCAGCCGAAU-------------------UCGA--------------UUUGAUUGAGACUCGGCUUGGAGUUUUUCCCCUGCUCAUACACUCACUUCAAGCCCGAGUCUCACGACU--AGGGCAAAUGCUCCCCGCAAUAAUAAA ((((.((((((....))))-------------------))))--------------))....((((((((((((((((((.....................)))))))).)))))))))).....--.((((....)))).............. ( -37.00, z-score = -3.23, R) >droGri2.scaffold_15074 6835339 119 - 7742996 GACCAGGCAUGAGCUGUCU-------------------UACUUGUC----------UCUGACU--GACUUUGUUUG----UCUUUCAUUAUUUAUUCUCUUAUCGCAAGCAAGGAUAACGUUUCCCCAAGGCAAAAACAAACGCAUACAGAGAA (((..(((....)))))).-------------------......((----------((((..(--(..((((((((----((((.......(((((((.....(....)..))))))).........))))))..))))))..))..)))))). ( -24.09, z-score = -0.37, R) >consensus GAUCAGAGUUGAGCCGAAU___________________UAGAUGUCUCUUUUUGG_UUUGAUUGAGACUCGGCUUGGAGUUUUUCCCCUGCUCAUACACUCACUUCAAGUCCGAGUCUCACGACC__AAGGCAAAUGCUACCU___A_____AA ..............................................................(((((((((((((((((.......................))))))))).)))))))).................................. (-15.30 = -16.14 + 0.84)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:46:42 2011