
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,440,207 – 15,440,347 |
| Length | 140 |
| Max. P | 0.887495 |

| Location | 15,440,207 – 15,440,314 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 70.93 |
| Shannon entropy | 0.52761 |
| G+C content | 0.42806 |
| Mean single sequence MFE | -21.20 |
| Consensus MFE | -9.26 |
| Energy contribution | -9.54 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.856291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
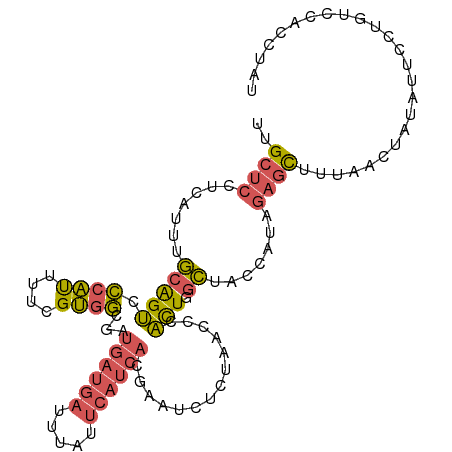
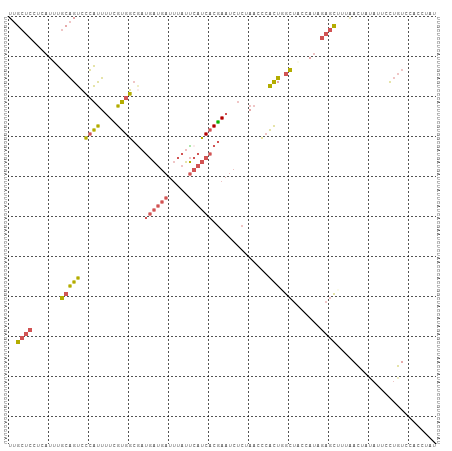
>dm3.chrX 15440207 107 - 22422827 UUGCUCCACAUUUGCAGUCCCAUUUUCGUGGCACUGAUGAUUUAUUCAUCACGAAUCUCUAACCCACUGGCUUCCAUAGAGCUUUAACUAUAUCCCUGUCCACCUAU ..((((.......(((((.((((....)))).)))(((((.....)))))...................)).......))))......................... ( -17.64, z-score = -1.27, R) >droSim1.chrX 11905370 107 - 17042790 UUGCUCCCCAUUUGCAGUCCCAUUUUCGUGGCGCUGAUGAUUUAUUCAUCACGAAUCUCUAACCCACUGGCUACCAUAGAGCUUUAACUCUAUCCCUGUCCAACUAU ((((.........))))..........((((((.((((((.....))))))))..........))))((((....((((((......))))))....).)))..... ( -20.20, z-score = -2.03, R) >droYak2.chrX 9624982 107 - 21770863 UUGCUCCUCAUUUGCAGUCCCAUUUUCGUGGCGAUGAUGAUUUAUUCAUCACGAAUCUCUAACCCACUUGCUGCCAUUGAGCUUUUACUACAUUCUUGUUCACCUUU ......((((...(((((.((((....))))((.((((((.....))))))))................)))))...)))).......................... ( -20.30, z-score = -2.37, R) >droEre2.scaffold_4690 7153801 107 + 18748788 UUGCUCCUCAUUUGCAGUCCCAUUUUUGUGGCGAUGAUGAUUUAUUCAUCACGAAUCUCUAACCCACUUGCUUCCACAGAGCUUUGACUAUAUUCCUGUCCACCUAU ..((((.......((((((((((....)))).)))(((((.....)))))..................))).......))))...(((.........)))....... ( -20.34, z-score = -2.14, R) >dp4.chrXL_group1a 1024456 100 - 9151740 UAGCGGGGGAUUGUCGGGGAACCUGUUGGGUGGAGCGAAACAAAUCAAACGCAA---UCUAAGGAUUUAAGGAUCAUC---UUUUAAGUUUGUUUUGUUCCAGCUU- (((((((..............)))))))(.(((((((((((((((.......((---((....)))).((((.....)---)))...))))))))))))))).)..- ( -27.54, z-score = -1.52, R) >consensus UUGCUCCUCAUUUGCAGUCCCAUUUUCGUGGCGAUGAUGAUUUAUUCAUCACGAAUCUCUAACCCACUGGCUACCAUAGAGCUUUAACUAUAUUCCUGUCCACCUAU ..((((.......(((((.((((....))))...((((((.....))))))..............))).)).......))))......................... ( -9.26 = -9.54 + 0.28)

| Location | 15,440,247 – 15,440,347 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 70.09 |
| Shannon entropy | 0.52353 |
| G+C content | 0.46160 |
| Mean single sequence MFE | -27.13 |
| Consensus MFE | -8.21 |
| Energy contribution | -8.97 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.887495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
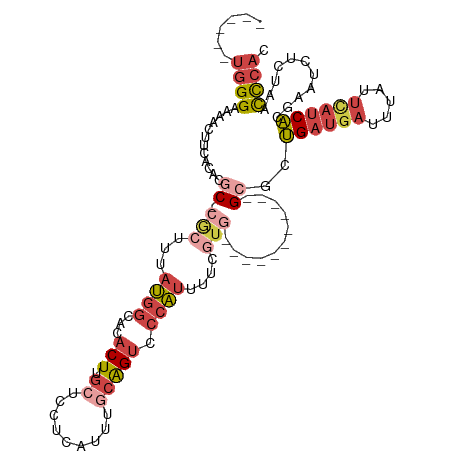
>dm3.chrX 15440247 100 - 22422827 ------UGGGAAAACUUCACACGC-CGCUUUAUGGCACACUUGCUCCACAUUUGCAGUCCCAUUUUCGUG-----------GCACUGAUGAUUUAUUCAUCACGAAUCUCUAACCCAC ------((((............((-(((...((((.((...(((.........))))).))))....)))-----------))..((((((.....))))))...........)))). ( -26.30, z-score = -3.68, R) >droSim1.chrX 11905410 100 - 17042790 ------UGGGAAAACUUCACACGC-CGCUUUAUGGCACACUUGCUCCCCAUUUGCAGUCCCAUUUUCGUG-----------GCGCUGAUGAUUUAUUCAUCACGAAUCUCUAACCCAC ------((((...........(((-(((...((((.((...(((.........))))).))))....)))-----------))).((((((.....))))))...........)))). ( -27.60, z-score = -3.54, R) >droYak2.chrX 9625022 100 - 21770863 ------UGGGAAAACUUCACACGC-CGCUUUAUGGCACACUUGCUCCUCAUUUGCAGUCCCAUUUUCGUG-----------GCGAUGAUGAUUUAUUCAUCACGAAUCUCUAACCCAC ------((((...........(((-(((...((((.((...(((.........))))).))))....)))-----------))).((((((.....))))))...........)))). ( -28.50, z-score = -3.92, R) >droEre2.scaffold_4690 7153841 100 + 18748788 ------UGGGAAAACUUCACACGC-CGCUUUAUGGCACACUUGCUCCUCAUUUGCAGUCCCAUUUUUGUG-----------GCGAUGAUGAUUUAUUCAUCACGAAUCUCUAACCCAC ------((((...........(((-(((...((((.((...(((.........))))).))))....)))-----------))).((((((.....))))))...........)))). ( -29.20, z-score = -4.11, R) >droAna3.scaffold_12929 430401 118 + 3277472 GACCGUUGGGAAAACUUUGCACACAAAGUUUAUGGUACACUUGGUCCUCAUUUGGAGUCCCAUUUUUGUAUUUUUGUGGUUGCUCUGAUGAUUUAUUUAUCACGUUUCUCUGACCCAC ....((..(..((((...((((((((((......(((((..(((..(((.....)))..)))....))))))))))))..)))..((((((.....)))))).))))..)..)).... ( -29.10, z-score = -1.52, R) >dp4.chrXL_group1a 1024492 94 - 9151740 ------UGGGGAAGGUCCUCUAGCAAA----AGGACGCACUAGCGGGGGAUUGUCGGGGAACCUGUUGGG----------UGGAGCGA-AACAAAUCAAACGCAAUCUAAGGAUU--- ------......((((.(((..((((.----....(((....))).....))))..))).)))).(((((----------(...(((.-...........))).)))))).....--- ( -22.10, z-score = 0.40, R) >consensus ______UGGGAAAACUUCACACGC_CGCUUUAUGGCACACUUGCUCCUCAUUUGCAGUCCCAUUUUCGUG___________GCGCUGAUGAUUUAUUCAUCACGAAUCUCUAACCCAC ......((((.........((((........((((...(((.((.........))))).))))...))))...............((((((.....))))))...........)))). ( -8.21 = -8.97 + 0.76)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:46:41 2011