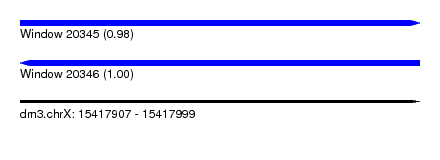
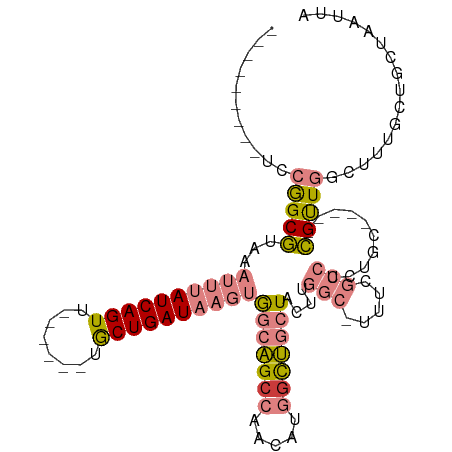
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,417,907 – 15,417,999 |
| Length | 92 |
| Max. P | 0.999648 |

| Location | 15,417,907 – 15,417,999 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 72.27 |
| Shannon entropy | 0.45624 |
| G+C content | 0.46005 |
| Mean single sequence MFE | -28.07 |
| Consensus MFE | -11.97 |
| Energy contribution | -12.66 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.983208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15417907 92 + 22422827 UAAUUAGCAGAAAAGCCAACG-----GCAGAGGCAAAAAGCCAAGUAGCAGCCAUGUUGGCUGCCACUUAUCAGUA------AACUGAUAAAUUUACGCCGGA--------- ......((......))...((-----((...(((.....)))..(((((((((.....))))))...(((((((..------..)))))))...)))))))..--------- ( -32.70, z-score = -4.40, R) >droPer1.super_26 1111897 81 + 1349181 UAAUUAGCAGCAAAGGCAGCC-----CCAGA-----------AAAUAGAGACAACUCGAGCCGUUUCUUAUCAGCA------AACUGAUUGCUUCUGGCCCGA--------- ......((.((....)).)).-----(((((-----------(...((((((..........)))))).(((((..------..)))))...)))))).....--------- ( -16.20, z-score = 0.02, R) >dp4.chrXL_group1a 996660 81 + 9151740 UAAUUAGCAGCAAAGGCAGCC-----CCAGA-----------AAAUAGAGACAACUCGAGCCGUUUCUUAUCAGCA------AACUGAUUGCUUCUGGCCCGA--------- ......((.((....)).)).-----(((((-----------(...((((((..........)))))).(((((..------..)))))...)))))).....--------- ( -16.20, z-score = 0.02, R) >droEre2.scaffold_4690 7131832 89 - 18748788 UAAUUAGCA-UAAAUCCAGCG-----GCAGAGGCGAAU-GCCAAGAAGCAGCCAUGUUGGCUGCCACUUAUCAGCA------AACUGAUAAAUUUACGCCGG---------- .........-.........((-----((...(((....-))).....((((((.....))))))...(((((((..------..)))))))......)))).---------- ( -29.00, z-score = -2.84, R) >droYak2.chrX 9602482 111 + 21770863 UAAUUAGCAGCAAAGCCAACGCAAAAGCAGAGGCAAAA-GCCAAGUAGCAGCCAUGUUGCCUGCCACUUAUCAGCACUUAAAAACUGAUAAAUUUACGCUGCCUUCUCACUG ......(((((...((....))....((((.(((....-)))..((((((....))))))))))...(((((((..........)))))))......))))).......... ( -29.60, z-score = -2.01, R) >droSec1.super_73 152336 91 + 167737 UAAUUAGCAGAAAAGCCAACG-----GCAGAGGCGAAA-GCCAAGUAGCAGCCAUGUUGGCUGCCACUUAUCAGUA------AACUGAUAAAUUUACGCCGGA--------- ......((......))...((-----((...(((....-)))..(((((((((.....))))))...(((((((..------..)))))))...)))))))..--------- ( -36.40, z-score = -5.57, R) >droSim1.chrX 11910239 91 + 17042790 UAAUUAGCAGAAAAGCCAACG-----GCAGAGGCGAAA-GCCAAGUAGCAGCCAUGUUGGCUGCCACUUAUCAGUA------AACUGAUAAAUUUACGCCGGA--------- ......((......))...((-----((...(((....-)))..(((((((((.....))))))...(((((((..------..)))))))...)))))))..--------- ( -36.40, z-score = -5.57, R) >consensus UAAUUAGCAGAAAAGCCAACG_____GCAGAGGCGAAA_GCCAAGUAGCAGCCAUGUUGGCUGCCACUUAUCAGCA______AACUGAUAAAUUUACGCCGGA_________ ......((.......................(((.....))).....((((((.....))))))...(((((((..........)))))))......))............. (-11.97 = -12.66 + 0.69)

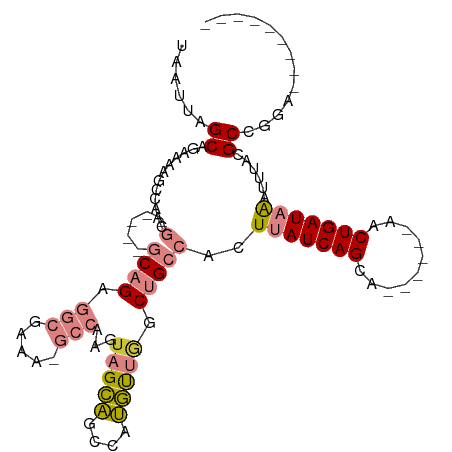
| Location | 15,417,907 – 15,417,999 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 72.27 |
| Shannon entropy | 0.45624 |
| G+C content | 0.46005 |
| Mean single sequence MFE | -34.56 |
| Consensus MFE | -20.99 |
| Energy contribution | -21.59 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.32 |
| Mean z-score | -3.70 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.13 |
| SVM RNA-class probability | 0.999648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15417907 92 - 22422827 ---------UCCGGCGUAAAUUUAUCAGUU------UACUGAUAAGUGGCAGCCAACAUGGCUGCUACUUGGCUUUUUGCCUCUGC-----CGUUGGCUUUUCUGCUAAUUA ---------..((((......(((((((..------..)))))))(((((((((.....)))))))))..(((.....)))...))-----))(((((......)))))... ( -39.20, z-score = -6.02, R) >droPer1.super_26 1111897 81 - 1349181 ---------UCGGGCCAGAAGCAAUCAGUU------UGCUGAUAAGAAACGGCUCGAGUUGUCUCUAUUU-----------UCUGG-----GGCUGCCUUUGCUGCUAAUUA ---------.....(((((((..(((((..------..))))).(((.(((((....))))).)))..))-----------)))))-----(((.((....)).)))..... ( -23.60, z-score = -0.68, R) >dp4.chrXL_group1a 996660 81 - 9151740 ---------UCGGGCCAGAAGCAAUCAGUU------UGCUGAUAAGAAACGGCUCGAGUUGUCUCUAUUU-----------UCUGG-----GGCUGCCUUUGCUGCUAAUUA ---------.....(((((((..(((((..------..))))).(((.(((((....))))).)))..))-----------)))))-----(((.((....)).)))..... ( -23.60, z-score = -0.68, R) >droEre2.scaffold_4690 7131832 89 + 18748788 ----------CCGGCGUAAAUUUAUCAGUU------UGCUGAUAAGUGGCAGCCAACAUGGCUGCUUCUUGGC-AUUCGCCUCUGC-----CGCUGGAUUUA-UGCUAAUUA ----------..((((((((((((((((..------..))))))((((((((((.....)))).......(((-....)))...))-----)))).))))))-))))..... ( -35.40, z-score = -3.83, R) >droYak2.chrX 9602482 111 - 21770863 CAGUGAGAAGGCAGCGUAAAUUUAUCAGUUUUUAAGUGCUGAUAAGUGGCAGGCAACAUGGCUGCUACUUGGC-UUUUGCCUCUGCUUUUGCGUUGGCUUUGCUGCUAAUUA ((((..((((.(((((((((.((((((((........))))))))(((((((.(.....).)))))))..(((-....)))......))))))))).))))))))....... ( -42.90, z-score = -2.94, R) >droSec1.super_73 152336 91 - 167737 ---------UCCGGCGUAAAUUUAUCAGUU------UACUGAUAAGUGGCAGCCAACAUGGCUGCUACUUGGC-UUUCGCCUCUGC-----CGUUGGCUUUUCUGCUAAUUA ---------..((((......(((((((..------..)))))))(((((((((.....)))))))))..(((-....)))...))-----))(((((......)))))... ( -38.60, z-score = -5.86, R) >droSim1.chrX 11910239 91 - 17042790 ---------UCCGGCGUAAAUUUAUCAGUU------UACUGAUAAGUGGCAGCCAACAUGGCUGCUACUUGGC-UUUCGCCUCUGC-----CGUUGGCUUUUCUGCUAAUUA ---------..((((......(((((((..------..)))))))(((((((((.....)))))))))..(((-....)))...))-----))(((((......)))))... ( -38.60, z-score = -5.86, R) >consensus _________UCCGGCGUAAAUUUAUCAGUU______UGCUGAUAAGUGGCAGCCAACAUGGCUGCUACUUGGC_UUUCGCCUCUGC_____CGUUGGCUUUGCUGCUAAUUA ...........(((((...((((((((((........))))))))))(((((((.....))))))).........................)))))................ (-20.99 = -21.59 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:46:36 2011