
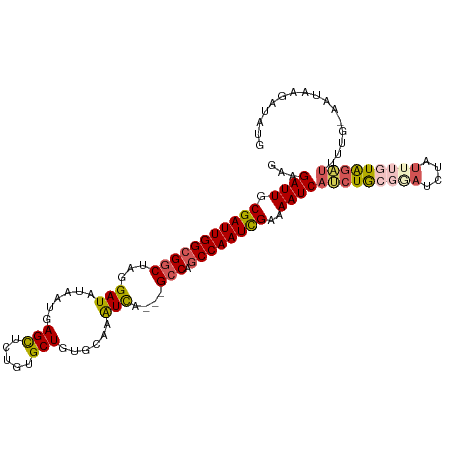
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,397,255 – 15,397,366 |
| Length | 111 |
| Max. P | 0.705065 |

| Location | 15,397,255 – 15,397,366 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 81.11 |
| Shannon entropy | 0.34675 |
| G+C content | 0.41936 |
| Mean single sequence MFE | -28.60 |
| Consensus MFE | -22.68 |
| Energy contribution | -23.64 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.611422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15397255 111 + 22422827 GAAGAUUGCGAUUGGCGGCUAGGAUAUAAUGAGUUGUGUGCUGUGCAAAUCGAAUGCCAACCAAUCGAAAAUCAUCUGCGAAUCUAUUUGUAAAUUUUUUAAUAAGACAUG ((.((((.(((((((.((((((.((((((....)))))).)))............)))..)))))))..)))).))((((((....))))))................... ( -22.40, z-score = 0.01, R) >droEre2.scaffold_4690 7111459 101 - 18748788 GAAGAUUGCGAUUGGCGACUAGGAUAUAAUGAGCUCUGUGCUGUGCAAGUCA---GCCAGCCAAUCGAAAAUCAGCUACGGAGCUAUGGAUAG-------GAUAGGAUACG .....((((.....))))(((...(((....((((((((((((.((......---)))))).....(.....)....))))))))....))).-------..)))...... ( -24.70, z-score = 0.17, R) >droYak2.chrX 9583032 101 + 21770863 GAAGAUUGCGAUUGGCGGCUAGGAUAUAACGAGCUCUGUGCUGUGCCAAUCA---GCCAGCCAAUUGAAAAUCAGCUA------UUUUAGUAGCUAUUC-AAUAAGAUAUG (((((((.((((((((((((..(((...((.(((.....)))))....))))---))).))))))))..))))(((((------(....)))))).)))-........... ( -31.10, z-score = -2.39, R) >droSec1.super_73 131666 110 + 167737 GAAGAUUGCGAUUGGCGGCUAGGAUAUAAUGAGCUCUGAGCUCUGCAAAUCGCCAGCCAGCCAAUCGAAAAUCAUCUGCGGAUCUAUUUGUGGAUUUUG-AAUAAGAUAUU ...((((.((((((((((((.((((.....((((.....)))).....))).).)))).))))))))..))))(((..((((....))))..)))....-........... ( -35.90, z-score = -2.33, R) >droSim1.chrX 11885467 110 + 17042790 GAAGAUUGCGAUUGGCGGCUAGGAUAUAAUGAGCUCUGUGCUGUGCAAAUUAGACGCCAGCCAAUCGAAAAUCAUCUGCGGAUCUAUUUGUGGAUUUUG-AAUAAGAUAUU ...((((.(((((((((((.......((((..((.(......).))..))))...))).))))))))..))))(((..((((....))))..)))....-........... ( -28.90, z-score = -0.94, R) >consensus GAAGAUUGCGAUUGGCGGCUAGGAUAUAAUGAGCUCUGUGCUGUGCAAAUCA___GCCAGCCAAUCGAAAAUCAUCUGCGGAUCUAUUUGUAGAUUUUG_AAUAAGAUAUG ...((((.(((((((((((...(((......(((.....)))......)))....))).))))))))..))))(((((((((....)))))))))................ (-22.68 = -23.64 + 0.96)

| Location | 15,397,255 – 15,397,366 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 81.11 |
| Shannon entropy | 0.34675 |
| G+C content | 0.41936 |
| Mean single sequence MFE | -24.29 |
| Consensus MFE | -18.30 |
| Energy contribution | -18.78 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.705065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15397255 111 - 22422827 CAUGUCUUAUUAAAAAAUUUACAAAUAGAUUCGCAGAUGAUUUUCGAUUGGUUGGCAUUCGAUUUGCACAGCACACAACUCAUUAUAUCCUAGCCGCCAAUCGCAAUCUUC ..........................(((((.((.((......))(((((((.(((....(((......((.......))......)))...))))))))))))))))).. ( -18.70, z-score = -0.63, R) >droEre2.scaffold_4690 7111459 101 + 18748788 CGUAUCCUAUC-------CUAUCCAUAGCUCCGUAGCUGAUUUUCGAUUGGCUGGC---UGACUUGCACAGCACAGAGCUCAUUAUAUCCUAGUCGCCAAUCGCAAUCUUC ...........-------........((((....))))((((..((((((((.(((---((........(((.....)))..........))))))))))))).))))... ( -22.17, z-score = -0.64, R) >droYak2.chrX 9583032 101 - 21770863 CAUAUCUUAUU-GAAUAGCUACUAAAA------UAGCUGAUUUUCAAUUGGCUGGC---UGAUUGGCACAGCACAGAGCUCGUUAUAUCCUAGCCGCCAAUCGCAAUCUUC .....((.(((-(((((((((......------))))))...)))))).))...((---.(((((((..(((.....))).((((.....)))).)))))))))....... ( -24.50, z-score = -1.17, R) >droSec1.super_73 131666 110 - 167737 AAUAUCUUAUU-CAAAAUCCACAAAUAGAUCCGCAGAUGAUUUUCGAUUGGCUGGCUGGCGAUUUGCAGAGCUCAGAGCUCAUUAUAUCCUAGCCGCCAAUCGCAAUCUUC ...........-..............((((..((.((......))(((((((.((((((.(((.....((((.....)))).....)))))))))))))))))).)))).. ( -33.90, z-score = -3.06, R) >droSim1.chrX 11885467 110 - 17042790 AAUAUCUUAUU-CAAAAUCCACAAAUAGAUCCGCAGAUGAUUUUCGAUUGGCUGGCGUCUAAUUUGCACAGCACAGAGCUCAUUAUAUCCUAGCCGCCAAUCGCAAUCUUC ...........-..............((((..((.((......))(((((((.((((((......).))(((.....)))............)))))))))))).)))).. ( -22.20, z-score = -1.21, R) >consensus CAUAUCUUAUU_CAAAAUCUACAAAUAGAUCCGCAGAUGAUUUUCGAUUGGCUGGC___UGAUUUGCACAGCACAGAGCUCAUUAUAUCCUAGCCGCCAAUCGCAAUCUUC ......................................((((..((((((((.(((....(((......(((.....)))......)))...))))))))))).))))... (-18.30 = -18.78 + 0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:46:33 2011