
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,397,109 – 15,397,203 |
| Length | 94 |
| Max. P | 0.847417 |

| Location | 15,397,109 – 15,397,203 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 64.38 |
| Shannon entropy | 0.67623 |
| G+C content | 0.59183 |
| Mean single sequence MFE | -35.76 |
| Consensus MFE | -11.87 |
| Energy contribution | -13.44 |
| Covariance contribution | 1.58 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.847417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
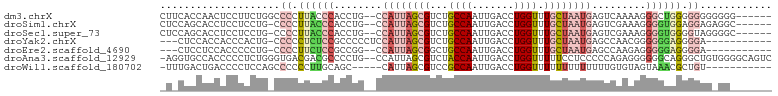
>dm3.chrX 15397109 94 + 22422827 CUUCACCAACUCCUUCUGGCCCCUUACCCACCUG--CCAUUAGCGUCUGCCAAUUGACCUGGUUUGCUAAUGAGUCAAAAGGGCUGGGGGGGGGGG------ .....(((........)))(((((..((((((((--(((((((((...((((.......)))).)))))))).)).....))).))))..))))).------ ( -37.20, z-score = -1.06, R) >droSim1.chrX 11885323 93 + 17042790 CUCCAGCACCUCCUCCUG-CCCCUUACCCACCUG--CCAUUAGCGUCUGCCAAUUGACCUGGUUUGCUAAUGAGUCGAAAGGGGUGGAGGAGAGGC------ .....((..(((((((.(-((((((.(......(--(((((((((...((((.......)))).)))))))).)).).))))))))))))))..))------ ( -40.80, z-score = -3.78, R) >droSec1.super_73 131522 93 + 167737 CUCCAGCACCUCCUCCUG-CCCCUUACCCACCUG--CCAUUAGCGUCUGCCAAUUGACCUGGUUUGCUAAUGAGUCGAAAGGGGUGGGGUAGGGGC------ .................(-(((((..((((((((--(((((((((...((((.......)))).)))))))).))(....))))))))..))))))------ ( -41.50, z-score = -2.84, R) >droYak2.chrX 9582894 87 + 21770863 ---CUCCACCACCCACUG-CCCCCUCUCCGCCCCCUCCAUUAGCGUCUGCCAAUUGACCUGGUUUGCUAAUGAGCCAACGGGGGGAGGGGA----------- ---...............-.(((((((((.((..(((.(((((((...((((.......)))).)))))))))).....))))))))))).----------- ( -34.60, z-score = -1.99, R) >droEre2.scaffold_4690 7111315 85 - 18748788 ---CUCCUCCACCCCCUG-CCCCUUCUCCGCCGG--CCAUUAGCGGCUGCCAAUUGACCUGGUUUGCUAAUGAGCCAAGAGGGGGAGGGGA----------- ---.........(((((.-((((((((.....((--((((((((((..((((.......))))))))))))).))).))))))))))))).----------- ( -42.10, z-score = -3.00, R) >droAna3.scaffold_12929 372676 99 - 3277472 -AGGUGCCACCCCCUCUGGGUGACGACGCCCCUG--CCAUUAGCGUCUACCAAUUGACCUGGUUUUUCCUCCCCCAGAGGGGGGCAGGGCUGUGGGGCAGUC -......(((((.....)))))..((((((((.(--(((((((.(((........)))))))).....(((((((....))))).)))))...))))).))) ( -43.50, z-score = -0.47, R) >droWil1.scaffold_180702 2911928 85 - 4511350 -UUUGACUGACCCCUCCAGCCCCCCUUGCAGC-----CAUUAGCGUCCGCCAAUUGACCUGGUUUUUUUUUUUUUGUGUAGUAAACGCUGU----------- -.....(((.......)))........(((((-----.(((((.(((........))))))))............((.......)))))))----------- ( -10.60, z-score = 0.54, R) >consensus _U_CACCACCUCCCCCUG_CCCCUUACCCACCUG__CCAUUAGCGUCUGCCAAUUGACCUGGUUUGCUAAUGAGUCAAAAGGGGUAGGGGAG_GG_______ ....................((.((((((........((((((((...((((.......)))).)))))))).........)))))).))............ (-11.87 = -13.44 + 1.58)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:46:32 2011