
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,377,592 – 15,377,667 |
| Length | 75 |
| Max. P | 0.877365 |

| Location | 15,377,592 – 15,377,667 |
|---|---|
| Length | 75 |
| Sequences | 7 |
| Columns | 85 |
| Reading direction | forward |
| Mean pairwise identity | 59.96 |
| Shannon entropy | 0.76457 |
| G+C content | 0.60822 |
| Mean single sequence MFE | -26.59 |
| Consensus MFE | -11.25 |
| Energy contribution | -13.31 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.877365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
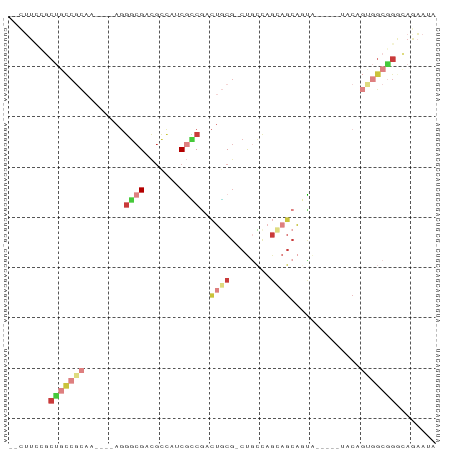
>dm3.chrX 15377592 75 + 22422827 --CUUCCGCUGCCGCAA----AGGGCGACGCCAUCGCCGACUGCG-CUGCCAGCAGCAGCAC---UUACAGAGGCAGGCAAAAUA --.....(((((((((.----..(((((.....)))))...)))(-((((.....)))))..---.......)))).))...... ( -29.00, z-score = -1.21, R) >droSec1.super_73 86481 78 + 167737 --CUUCCGCUGCCGCAA----AGGGCGACGCCAUCGCCGACUGCG-CUGCCAGCAGCAGCAGCACUUACAGUGGCGGGCAGAAUA --.(((.((((((((..----..(((((.....)))))(.(((((-(((....)))).))))).......))))).))).))).. ( -34.00, z-score = -1.30, R) >droYak2.chrX 9563168 72 + 21770863 --CUUCCGCUGCCGCAA----AGGGCGACGUCAACGUCGACUGCG-CUGCCAGCAGCACUG------ACAGUGGCGGACAGAAUG --(((((((..(.(((.----....(((((....)))))..)))(-(((....))))....------...)..))))).)).... ( -28.90, z-score = -1.32, R) >droEre2.scaffold_4690 7092497 72 - 18748788 --CUUUUGCUGCCGCAA----AGGGCGACGCCAACGCCGACUGCG-CUGCUAGAAGCAGUG------ACAGUGGCGGGUAGAAUA --......(((((....----..(((...)))..((((.((((((-((((.....))))))------.))))))))))))).... ( -32.80, z-score = -2.83, R) >droSim1.chrX_random 4016260 78 + 5698898 --CUUCCGCUGCCGCAA----AGGGCGACGCCAUCGCCGACUGCG-CUGCCAGCAGCAGCAGCACUUACAGUGGCGGGCAGAAUA --.(((.((((((((..----..(((((.....)))))(.(((((-(((....)))).))))).......))))).))).))).. ( -34.00, z-score = -1.30, R) >droAna3.scaffold_12929 348271 83 - 3277472 UUUAUGCUCCAUCAUUAUGCCAUGCCGAGCUGUGCAGCUGUUCCUUCCCGCAUCCGCACUAUUG--UACACUUGCGGACAAUGCG .....((((...(((......)))..))))...(((..(((......(((((...(.((....)--).)...)))))))).))). ( -15.90, z-score = 0.62, R) >droWil1.scaffold_180702 2890246 58 - 4511350 --CUUCCUCAGGCACAACAUCAGUGCGAGGCAAGCAGCAGCAUCG-CCAGCAGACUCUAUC------------------------ --...((((..((((.......))))))))...((.((......)-)..))..........------------------------ ( -11.50, z-score = 0.11, R) >consensus __CUUCCGCUGCCGCAA____AGGGCGACGCCAUCGCCGACUGCG_CUGCCAGCAGCAGUA_____UACAGUGGCGGGCAGAAUA ........(((((((........((((.......))))..((((........))))..............)))))))........ (-11.25 = -13.31 + 2.06)


| Location | 15,377,592 – 15,377,667 |
|---|---|
| Length | 75 |
| Sequences | 7 |
| Columns | 85 |
| Reading direction | reverse |
| Mean pairwise identity | 59.96 |
| Shannon entropy | 0.76457 |
| G+C content | 0.60822 |
| Mean single sequence MFE | -29.83 |
| Consensus MFE | -10.40 |
| Energy contribution | -11.39 |
| Covariance contribution | 0.99 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.814480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
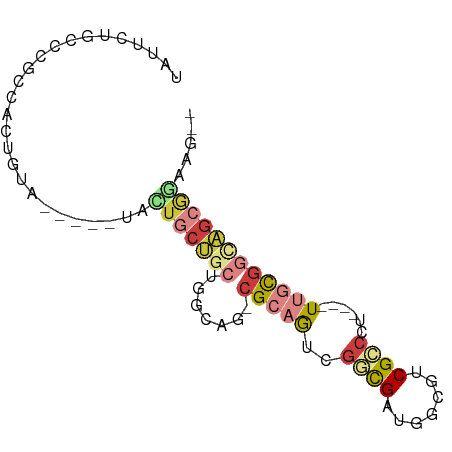
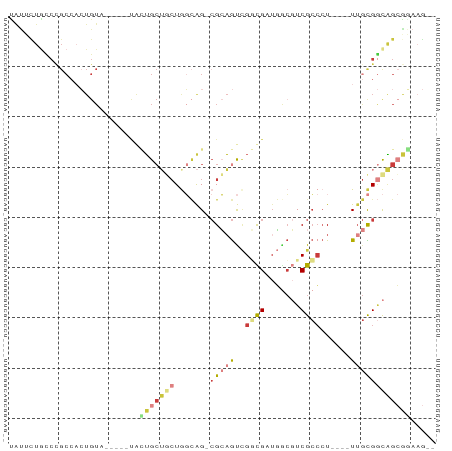
>dm3.chrX 15377592 75 - 22422827 UAUUUUGCCUGCCUCUGUAA---GUGCUGCUGCUGGCAG-CGCAGUCGGCGAUGGCGUCGCCCU----UUGCGGCAGCGGAAG-- ..((((((.((((.......---..(((((.....))))-)((((..((((((...))))))..----)))))))))))))).-- ( -34.70, z-score = -1.53, R) >droSec1.super_73 86481 78 - 167737 UAUUCUGCCCGCCACUGUAAGUGCUGCUGCUGCUGGCAG-CGCAGUCGGCGAUGGCGUCGCCCU----UUGCGGCAGCGGAAG-- ..((((((..(((.........((((((......)))))-)((((..((((((...))))))..----))))))).)))))).-- ( -37.00, z-score = -1.33, R) >droYak2.chrX 9563168 72 - 21770863 CAUUCUGUCCGCCACUGU------CAGUGCUGCUGGCAG-CGCAGUCGACGUUGACGUCGCCCU----UUGCGGCAGCGGAAG-- ..((((((..(((.((((------(((.....)))))))-.((((.(((((....)))))....----))))))).)))))).-- ( -31.90, z-score = -1.93, R) >droEre2.scaffold_4690 7092497 72 + 18748788 UAUUCUACCCGCCACUGU------CACUGCUUCUAGCAG-CGCAGUCGGCGUUGGCGUCGCCCU----UUGCGGCAGCAAAAG-- .........((((((((.------(.((((.....))))-.))))).))))...(((((((...----..))))).)).....-- ( -27.10, z-score = -2.12, R) >droSim1.chrX_random 4016260 78 - 5698898 UAUUCUGCCCGCCACUGUAAGUGCUGCUGCUGCUGGCAG-CGCAGUCGGCGAUGGCGUCGCCCU----UUGCGGCAGCGGAAG-- ..((((((..(((.........((((((......)))))-)((((..((((((...))))))..----))))))).)))))).-- ( -37.00, z-score = -1.33, R) >droAna3.scaffold_12929 348271 83 + 3277472 CGCAUUGUCCGCAAGUGUA--CAAUAGUGCGGAUGCGGGAAGGAACAGCUGCACAGCUCGGCAUGGCAUAAUGAUGGAGCAUAAA .((((((((((((..((((--(....)))))..)))))......)))).)))...((((.((((......))).).))))..... ( -25.90, z-score = -1.01, R) >droWil1.scaffold_180702 2890246 58 + 4511350 ------------------------GAUAGAGUCUGCUGG-CGAUGCUGCUGCUUGCCUCGCACUGAUGUUGUGCCUGAGGAAG-- ------------------------....((((..((.((-(...))))).)))).((((((((.......))))..))))...-- ( -15.20, z-score = 0.13, R) >consensus UAUUCUGCCCGCCACUGUA_____UACUGCUGCUGGCAG_CGCAGUCGGCGAUGGCGUCGCCCU____UUGCGGCAGCGGAAG__ ..........................(((((((((((....))....((((.......)))).........)))))))))..... (-10.40 = -11.39 + 0.99)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:46:27 2011