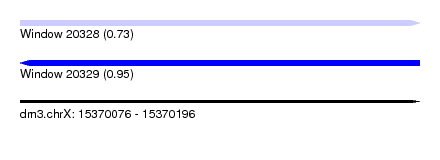
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,370,076 – 15,370,196 |
| Length | 120 |
| Max. P | 0.945136 |

| Location | 15,370,076 – 15,370,196 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.03 |
| Shannon entropy | 0.33419 |
| G+C content | 0.48319 |
| Mean single sequence MFE | -34.75 |
| Consensus MFE | -19.86 |
| Energy contribution | -22.79 |
| Covariance contribution | 2.94 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.725297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15370076 120 + 22422827 AAGUGGCAUCGAUAUCGUAAUGUCCUACGUAUCGAUGCUGCACCAUUCAUUAACGGCCCAACGCUUGUCAAGGCAACAUUGUCUCUUUGGUAAGUCUCCCGGCAUCCGUUUCCGGAAAAU ..(..(((((((((.((((......)))))))))))))..)..............(((....((((..(((((((....)))))...))..)))).....))).((((....)))).... ( -39.30, z-score = -3.88, R) >droSec1.super_73 79120 119 + 167737 -AGUGGCAUCGAUAUCGGAAUGUCCUACGUAUCAAUGCUACACCGUUCAUUAACGGCCCAACGCUCGAGGACGCAACAUUGUCUCUUUGGUAAGUCUCCCAGCAUCCGUUUCCGGAAAAG -.(((((((.(((((.((.....))...))))).))))))).(((((....)))))......(((.(..(((....((.........))....)))..).))).((((....)))).... ( -32.80, z-score = -1.60, R) >droYak2.chrX 9558310 120 + 21770863 UGAAGCCAUCGAUAUCCUGAUGUCCUACGUAUCGAUGCUGCACUGCCAUUUAACGGCUCAUCGCUUAAGGAGACGUCAUUUUACCUUUGGCAAGUGGACUGGCAUCCGUUUCCGGAAAAU ....(((((((((((...(.....)...))))))))....(((((((.......))).....(((.(((((((......))).)))).))).))))....))).((((....)))).... ( -31.00, z-score = -0.41, R) >droEre2.scaffold_4690 7086768 115 - 18748788 UGAAGCCAUCGAUAUCGUGAUGUCCUACGUAUCGAUGCUGCACUGCCAUUUAACGGUCCAUCCCUCGAGGA-----CAUUUUACCUUUGGUAACUGGACCGGCAUCCGUUUCCGGAAAAU ((.(((.(((((((.((((......)))))))))))))).)).((((.......(((((((..((.((((.-----.......)))).))..).))))))))))((((....)))).... ( -35.91, z-score = -2.38, R) >consensus UAAAGCCAUCGAUAUCGUAAUGUCCUACGUAUCGAUGCUGCACCGCCAAUUAACGGCCCAACGCUCGAGGAGGCAACAUUGUACCUUUGGUAAGUCGACCGGCAUCCGUUUCCGGAAAAU ..((((((((((((.((((......))))))))))))))))............(((.(((..(((.((((.............)))).)))...))).)))...((((....)))).... (-19.86 = -22.79 + 2.94)

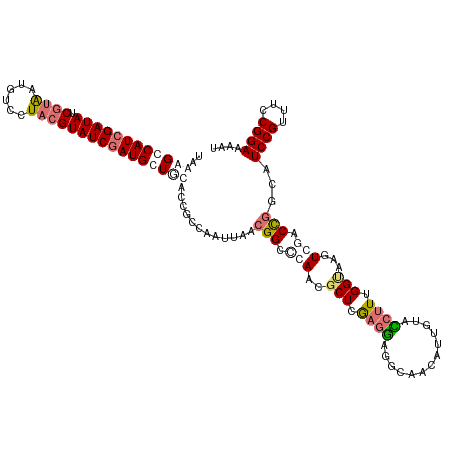

| Location | 15,370,076 – 15,370,196 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.03 |
| Shannon entropy | 0.33419 |
| G+C content | 0.48319 |
| Mean single sequence MFE | -38.86 |
| Consensus MFE | -25.06 |
| Energy contribution | -26.12 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.945136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15370076 120 - 22422827 AUUUUCCGGAAACGGAUGCCGGGAGACUUACCAAAGAGACAAUGUUGCCUUGACAAGCGUUGGGCCGUUAAUGAAUGGUGCAGCAUCGAUACGUAGGACAUUACGAUAUCGAUGCCACUU ....((((....))))(((.((........)).......(((((((.........))))))).((((((....)))))))))((((((((((((((....))))).)))))))))..... ( -40.40, z-score = -3.29, R) >droSec1.super_73 79120 119 - 167737 CUUUUCCGGAAACGGAUGCUGGGAGACUUACCAAAGAGACAAUGUUGCGUCCUCGAGCGUUGGGCCGUUAAUGAACGGUGUAGCAUUGAUACGUAGGACAUUCCGAUAUCGAUGCCACU- ((((((((....))))...(((........))).)))).(((((((.((....))))))))).((((((....))))))((.(((((((((....((.....))..))))))))).)).- ( -39.30, z-score = -2.13, R) >droYak2.chrX 9558310 120 - 21770863 AUUUUCCGGAAACGGAUGCCAGUCCACUUGCCAAAGGUAAAAUGACGUCUCCUUAAGCGAUGAGCCGUUAAAUGGCAGUGCAGCAUCGAUACGUAGGACAUCAGGAUAUCGAUGGCUUCA ....((((....)))).((((((((..(((((...)))))..(((.((((.......(((((.(((((...))))).......))))).......)))).))))))).....)))).... ( -35.34, z-score = -1.26, R) >droEre2.scaffold_4690 7086768 115 + 18748788 AUUUUCCGGAAACGGAUGCCGGUCCAGUUACCAAAGGUAAAAUG-----UCCUCGAGGGAUGGACCGUUAAAUGGCAGUGCAGCAUCGAUACGUAGGACAUCACGAUAUCGAUGGCUUCA ....((((....))))(((((((((..(((((...)))))...(-----(((.....))))))))).......)))).((.(((((((((((((........))).))))))).))).)) ( -40.41, z-score = -2.67, R) >consensus AUUUUCCGGAAACGGAUGCCGGGACACUUACCAAAGAGAAAAUGUUGCCUCCUCAAGCGAUGGGCCGUUAAAGAACAGUGCAGCAUCGAUACGUAGGACAUCACGAUAUCGAUGCCACCA ....((((....)))).(((......................................((((.((((((....)))))).))))((((((((((((....))))).)))))))))).... (-25.06 = -26.12 + 1.06)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:46:22 2011