| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,355,680 – 15,355,800 |
| Length | 120 |
| Max. P | 0.500000 |

| Location | 15,355,680 – 15,355,800 |
|---|---|
| Length | 120 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.31 |
| Shannon entropy | 0.62392 |
| G+C content | 0.54646 |
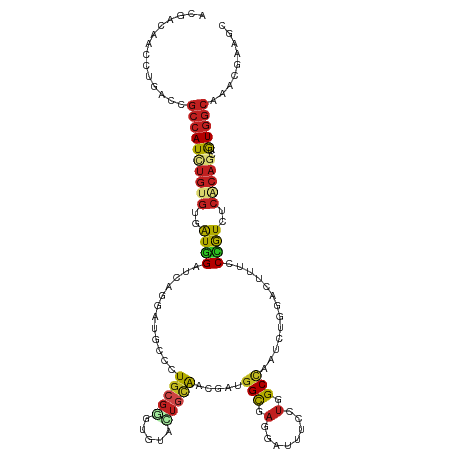
| Mean single sequence MFE | -41.50 |
| Consensus MFE | -17.49 |
| Energy contribution | -17.76 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
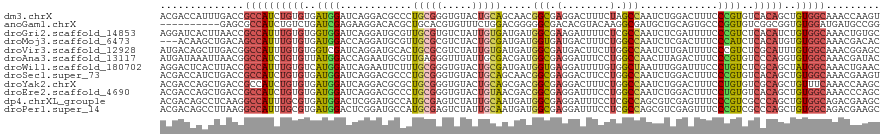
>dm3.chrX 15355680 120 - 22422827 ACGACCAUUUGACCGCCAUCUGUGUGAUGGAUCAGGACGCCCUGCGGGUGUACUGCAGCAACGGCGAGGACUUUCUAGCCAAUCUGGACUUUCCCGUGUCACAGCUGUGGCAAACCAAGU ....((.(((((...(((((.....))))).))))).((((((((((.....))))))....)))).))........((((..((((((........))).)))...))))......... ( -39.90, z-score = -0.40, R) >anoGam1.chrX 6860019 110 - 22145176 ----------GAGCGCCAUCUGUCUGAUCGAGAAGGACACGCUGCACGUGUUUCUGGACGGGGGCGACACGUACAAGGCGAUGCUGCAGUGCCCGGUGUCGGCGGUGUGGAUGAUGCCGG ----------..(((((.(((((((.....(((..((((((.....)))))))))))))))))))).)........((((...(..((.((((.......)))).))..)....)))).. ( -39.70, z-score = 1.04, R) >droGri2.scaffold_14853 10030132 120 + 10151454 AGGAUCACUUAACCGCCAUUUGUGUGGUGGAUCAGGAUGCGUUGCGUGUCUAUUGUGAUGAUGGCGAAGAUUUUCUCGCCAAUCUCGAUUUUCCCGUCUCACAUCUGUGGCAAACUGUGC ..(.((((....(((((((....)))))))....(((((.(..(((.(...((((.(((..((((((........))))))))).))))...).)))..).))))))))))......... ( -36.80, z-score = -1.11, R) >droMoj3.scaffold_6473 1168803 117 - 16943266 ---ACAAGCUGACAGCCAUUUGUGUGAUGGACCAGGAUGCGUUGCGCGUCUACUGCGAUGAUGGUGAUGACUUUCUGGCCAAUCUCGACUUUCCCAUCUCACAUGUGUGGCAAACGACAC ---...........(((((.((((.(((((....(((((((...)))))))....(((.((((((.(.(.....)).))).))))))......))))).))))...)))))......... ( -36.90, z-score = -0.99, R) >droVir3.scaffold_12928 3319211 120 - 7717345 AUGACAGCUUGACGGCCAUUUGUGUGGUCGAUCAGGAUGCACUGCGCGUCUAUUGUGAUGAUGGCGAUGACUUCUUGGCCAAUCUUGAUUUUCCCGUCUCGCAUUUGUGGCAAACGGAGC ......((((....(((((..((((((.((((((((((........((((......)))).((((.(........).)))))))))))).....)).).)))))..))))).....)))) ( -35.80, z-score = 0.19, R) >droAna3.scaffold_13117 1558123 120 - 5790199 AUGAUAAAUUAACGGCCAUCUGUGUUAUGGACCAGAAUGCGUUGAGGGUUUAUUGCGACGAUGGCGAGGAUUUCCUGGCCAACUUAGACUUUCCCGUGUCCCAGGUGUGGCAAACGAUAC ..((((((((..((((((((((.(((...)))))).))).))))..))))))))((.((..((((.(((....))).))))((((.(((........)))..)))))).))......... ( -35.00, z-score = -0.78, R) >droWil1.scaffold_180702 2883599 120 + 4511350 AGGACUCACUUACCGCCAUUUGUGUCAUGGAUCAGAAUUCUUUGCGGGUGUACUGCGAUGAUGGUGAGGAUUUUGUGGCUAAUUUGGAUUUCCCUGUCUCGCAGCUAUGGCAAACUGAAC ..............((((((((((.((.((((((((..((.((((((.....)))))).))((((.(.(....).).)))).)))))...))).))...)))))..)))))......... ( -27.10, z-score = 1.41, R) >droSec1.super_73 45415 120 - 167737 ACGACCAUCUGACCGCCAUCUGUGUGAUGGAUCAGGACGCCCUGCGGGUGUACUGCAGCAACGGCGAGGACUUCCUGGCCAAUCUGGACUUUCCCGUGUCACAGCUGUGGCAAACGAAGU ....((.(((((...(((((.....))))).))))).((((((((((.....))))))....)))).))(((((...((((..((((((........))).)))...))))....))))) ( -46.40, z-score = -1.47, R) >droYak2.chrX 9543858 120 - 21770863 ACGACCAGCUGACCGCCAUCUGUGUGAUGGAUCAGGACGCGCUGCGGGUGUACUGCAGCGACGGCGAGGACUUUCUGGCCAAUCUGGACUUUCCUGUGUCGCAGCUGUUUCAAACCAAGC ..((.((((((....(((((.....)))))..(((((.(((((((((.....)))))))).)(((.(((....))).)))...........))))).....))))))..))......... ( -46.00, z-score = -1.26, R) >droEre2.scaffold_4690 7071434 120 + 18748788 ACGACCAGCUGACCGCCAUCUGUGUGAUGGAUCAGGACGCCCUGCGGGUGUACUGUAACGACGGCGAGGAUUUCCUGGCCAAUCUGGACUUUCCUGUGUCACAGCUGUGGCAAACCCAGC .....((((((....(((((.....)))))..(((.((((((...)))))).)))....((((((.(((....))).))).....((.....))...))).))))))(((.....))).. ( -44.00, z-score = -1.06, R) >dp4.chrXL_group1e 12297287 120 - 12523060 ACGACAGCCUCAAGGCCAUUUGCGUGAUGGACUCGGAUGCCAUGCGAGUCUAUUGCAAUGAUGGCGAGGAUUUCCUCGCCAGCGUCGAGUUUCCCGUCGCCCAGCUGUGGCAGACGAAGC .((((.(((....))).....(((..(((((((((.((...)).)))))))))..).....((((((((....)))))))))))))).......((((((((....).))).)))).... ( -55.20, z-score = -4.06, R) >droPer1.super_14 858044 120 + 2168203 ACGACAGCCUUAAGGCCAUUUGCGUGAUGGACUCGGAUGCCAUGCGAGUCUAUUGCAAUGAUGGCGAGGAUUUCCUCGCCAGCGUCGAGUUUCCCGUCGCCCAGCUGUGGCAGACGAAGC .((((.(((....))).....(((..(((((((((.((...)).)))))))))..).....((((((((....)))))))))))))).......((((((((....).))).)))).... ( -55.20, z-score = -4.38, R) >consensus ACGACAACCUGACCGCCAUCUGUGUGAUGGAUCAGGAUGCCCUGCGGGUGUACUGCAACGAUGGCGAGGAUUUCCUGGCCAAUCUGGACUUUCCCGUCUCACAGCUGUGGCAAACGAAGC ..............((((((((((..((((............(((((.....))))).....(((.(........).))).............))))..)))))..)))))......... (-17.49 = -17.76 + 0.27)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:46:21 2011