
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,354,880 – 15,354,970 |
| Length | 90 |
| Max. P | 0.850432 |

| Location | 15,354,880 – 15,354,970 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 62.97 |
| Shannon entropy | 0.75892 |
| G+C content | 0.45011 |
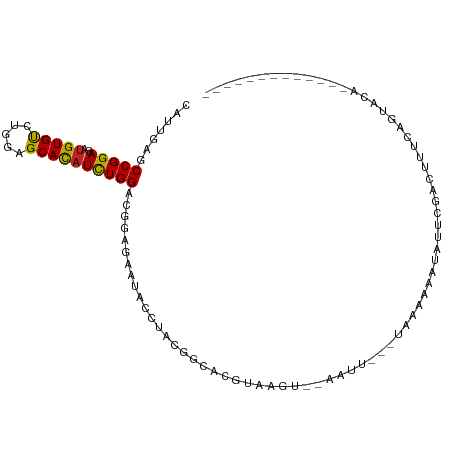
| Mean single sequence MFE | -23.35 |
| Consensus MFE | -9.70 |
| Energy contribution | -9.13 |
| Covariance contribution | -0.57 |
| Combinations/Pair | 1.30 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.850432 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15354880 90 - 22422827 CAUUGAGCCGGAAUUGUGUAUGGAGCACAUCUGGACGGAGAACAUCUACGGAACGUCAGU--AAAUG-CCACCAAAGAUGCCACUUUCAGUAC-------------- .(((((((((((..(((((.....))))))))))..((....(((((..((..(((....--..)))-...))..)))))))...))))))..-------------- ( -19.30, z-score = 0.25, R) >droSec1.super_73 44617 88 - 167737 CAUUGAGCCGGAAUUGUGCAUGGAGCACAUCUGGACGGAGAACACCUACGGAACGUCAGU--AAUU---UAUAAAAGAUGUCACUUUCAGUAC-------------- .(((((((((((..(((((.....))))))))))..((......))...(..(((((...--....---.......)))))..).))))))..-------------- ( -19.54, z-score = -0.63, R) >droYak2.chrX 9543057 90 - 21770863 CAUUGAACCGGAAUUGUGUCUGGAGCAUAUUUGGACGGAGAACACCUACGGAACGUCUGU--AAUC---UCAAAAAUAUGUGACUUUUUUUGUAC------------ .......(((((......))))).((((((((.....((((.....(((((.....))))--).))---))..))))))))..............------------ ( -18.90, z-score = -0.47, R) >droEre2.scaffold_4690 7070635 88 + 18748788 CAUUGAACCGGAAUUGUGUCUGGAGCACAUCUGGACGGAGAACAUUUACGGAACGUCUGU--AAUA---UUAGAAAGAUGUGAUUUUCAGUAC-------------- .(((((((((((......)))))..(((((((.....((......((((((.....))))--))..---))....)))))))...))))))..-------------- ( -22.20, z-score = -1.58, R) >droAna3.scaffold_13117 1557368 96 - 5790199 CAUCGAGCCGGAGAUGUGUCUGGAGCACCUCUGGACCGAAAACACCUACGGAACGUAAGAUACAUAUUCUUUAAGGCAUAUUGUAAUUUGUUAAUC----------- ..(((..((((((..((((.....))))))))))..))).(((((.((((...)))).).(((((((.((....)).))).))))...))))....----------- ( -19.40, z-score = 0.05, R) >dp4.chrXL_group1e 12296443 91 - 12523060 CAUCGAGCCGGAGAUGUGUCUGGAGCAUAUUUGGACAGAGAAUACCUAUGGCAGGUACGU--GAUUUAAUGUGUCCUGUUCGGCUUUCUUAAG-------------- ....(((((((...(.((((..((.....))..)))).)...........((((((((((--......))))).)))))))))))).......-------------- ( -28.50, z-score = -2.03, R) >droPer1.super_14 857203 92 + 2168203 CAUCGAGCCGGAGAUGUGUCUGGAGCAUAUUUGGACAGAGAAUACCUAUGGCAGGUACGU--GAUUUAAUUUUUCCUGUUCGGCUUUCUUUAAG------------- ..((((((.((((((.((((..((.....))..)))).)...(((((.....)))))...--.........))))).))))))...........------------- ( -24.50, z-score = -1.20, R) >droWil1.scaffold_180702 2882794 103 + 4511350 CAUUGAGCCGGAAAUGUGUUUGGAGCACAUUUGGACGGAGAAUUCAUACAGCCCGUAAGUUGAGUUACCUCCAAGUGAUUCAUCUCAUAAUCUCUCUCCCUCU---- ....(((....((((((((.....))))))))(((.(((((.....(((.....)))((.((((((((......)))))))).)).....))))).)))))).---- ( -26.60, z-score = -1.13, R) >droVir3.scaffold_12928 3318318 93 - 7717345 CAUUGAGCCGGAGAUGUGCUUGGAGCAUAUCUGGACAGAGAAUACGUACGGCACGUAAGUUUGUCC---GAAAACCAACAACAACUGCU-CCCAACU---------- ....((((.(.((((((((.....))))))))(((((((...(((((.....)))))..)))))))---...............).)))-)......---------- ( -28.40, z-score = -2.74, R) >droMoj3.scaffold_6473 1167951 102 - 16943266 CAUUGAGCCGGAGAUGUGUUUGGAGCACAUCUGGACAGAGAAUACCUAUGGCACGUAAGUU--CAC---AAAUAGCUCCUCAACCAGCCACUCAGCCACUCGACGCU ..(((((((..((((((((.....))))))))))..............((((((....)).--...---.....(((........)))......))))))))).... ( -22.70, z-score = 0.52, R) >droGri2.scaffold_14853 10029269 94 + 10151454 CAUUGAGCCGGAGAUGUGCUUGGAGCACAUCUGGACGGAGAAUACCUAUGGCACGUAAGUUGAUGU---GAAUGGUUACUCAAAC-CUAACCUAAUCU--------- ..((((((((.((((((((.....))))))))...))).......((((..(((((......))))---).))))...)))))..-............--------- ( -26.80, z-score = -1.91, R) >consensus CAUUGAGCCGGAGAUGUGUCUGGAGCACAUCUGGACGGAGAAUACCUACGGCACGUAAGU__AAUU___UAAAAAAUAUUCGACUUUCAGUACA_____________ .......(((((..(((((.....))))))))))......................................................................... ( -9.70 = -9.13 + -0.57)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:46:20 2011