| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,333,775 – 15,333,865 |
| Length | 90 |
| Max. P | 0.731431 |

| Location | 15,333,775 – 15,333,865 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 73.31 |
| Shannon entropy | 0.52397 |
| G+C content | 0.43921 |
| Mean single sequence MFE | -16.65 |
| Consensus MFE | -10.45 |
| Energy contribution | -10.19 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.24 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.731431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
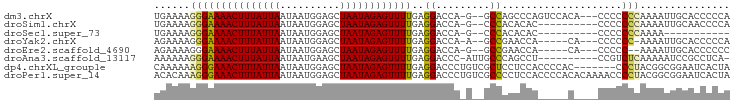
>dm3.chrX 15333775 90 + 22422827 UGAAAAGGGAAAACUUUAUUAAUAAUGGAGCUAAUAGAGUUUUGAGGACCA-G--GCCAGCCCAGUCCACA---CCCCCCCAAAAUUGCACCCCCA ......(((((((((((((((..........))))))))))))..((((..-(--(....))..))))...---....)))............... ( -22.20, z-score = -2.15, R) >droSim1.chrX 11848117 83 + 17042790 UGAAAAGGGAAAACUUUAUUAAUAAUGGAGCUAAUAGAGUUUUGAGGACCA-G--CCCACACAC----------CCCCCCCAAAAUUGCAACCCCA ......(((((((((((((((..........))))))))))))..((....-.--.))......----------....)))............... ( -14.70, z-score = -0.95, R) >droSec1.super_73 23838 72 + 167737 UGAAAAGGGAAAACUUUAUUAAUAAUGGAGCUAAUAGAGUUUUGAGGACCA-G--CCCACACAC----------CCCCCCCAAAA----------- ......(((((((((((((((..........))))))))))))..((....-.--.))......----------....)))....----------- ( -14.70, z-score = -1.68, R) >droYak2.chrX 9522645 84 + 21770863 AGAAAAGGGAAAACUUUAUUAAUAAUGGAGCUAAUAGAGUUUUGAGGACCA-A--GCCGAACCA-----CA---CCCCCC-AAAAUUGCACCCCCA ......(((((((((((((((..........))))))))))))..((....-.--.))......-----..---...)))-............... ( -14.50, z-score = -1.06, R) >droEre2.scaffold_4690 7050377 83 - 18748788 AGAAAAGGGAAAACUUUAUUAAUAAUGGAGCUAAUAGAGUUUUGAGGACCA-G--GCCGAACCA-----CA---CCCCC--AAAAUUGCACCCCCC ......(((((((((((((((..........))))))))))))..((....-.--.))......-----..---..)))--............... ( -14.50, z-score = -0.34, R) >droAna3.scaffold_13117 1538216 84 + 5790199 AAAAAAGGGAAAACUUUAUUAAUAAUGAAGCUAAUAGAGUUUUGAGGACCC-AUUGCCCAGCCU----------CCGUCUCAAAAAUCCGCCUCA- ......(((((((((((((((..........))))))))))))((((....-.........)))----------)...........)))......- ( -13.92, z-score = -0.29, R) >dp4.chrXL_group1e 12274698 89 + 12523060 CAAAAAAGGGAAACUUUAUUAAUAAUGGAGCUAAUAGAGUUUUGAGGACCCUGUCGCUCCUCCACCCCAC-------CCCUACGGCGGAAUCACUA .......((((((((((((((..........))))))))))).(((((.(.....).)))))...))).(-------(.(....).))........ ( -20.20, z-score = -0.64, R) >droPer1.super_14 835295 96 - 2168203 ACACAAAGGGAAACUUUAUUAAUAAUGGAGCUAAUAGAGUUUUGAGGACCCUGUCGCCCCUCCACCCCACACAAAACCCCUACGGCGGAAUCACUA .......((((((((((((((..........))))))))))).((((...........))))...)))........((.(....).))........ ( -18.50, z-score = -0.10, R) >consensus AGAAAAGGGAAAACUUUAUUAAUAAUGGAGCUAAUAGAGUUUUGAGGACCA_G__GCCACACCA_____C____CCCCCCCAAAAUUGCACCCCCA ......(((((((((((((((..........))))))))))))..((.........))....................)))............... (-10.45 = -10.19 + -0.27)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:46:16 2011