
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,305,183 – 15,305,323 |
| Length | 140 |
| Max. P | 0.898102 |

| Location | 15,305,183 – 15,305,323 |
|---|---|
| Length | 140 |
| Sequences | 6 |
| Columns | 168 |
| Reading direction | reverse |
| Mean pairwise identity | 67.74 |
| Shannon entropy | 0.55540 |
| G+C content | 0.44031 |
| Mean single sequence MFE | -39.84 |
| Consensus MFE | -16.76 |
| Energy contribution | -16.90 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.898102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
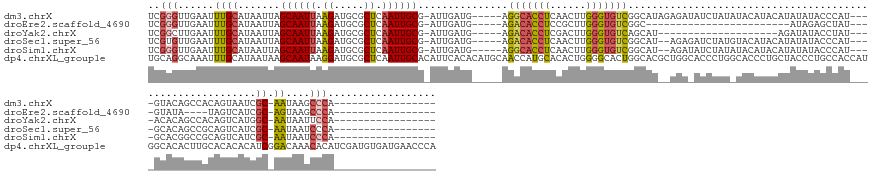
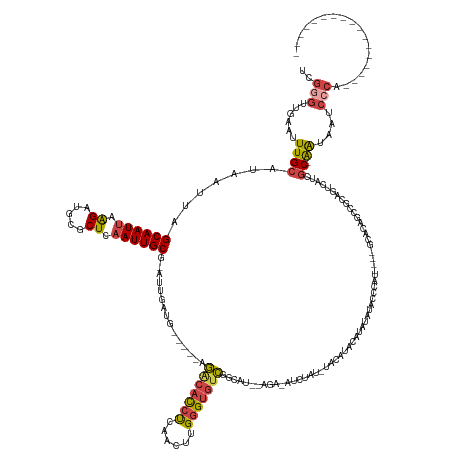
>dm3.chrX 15305183 140 - 22422827 UCGGGUUGAAUUUGCAUAAUUAGCAAUUAAGAUGCGCUCAAUUGCG-AUUGAUG-----AGGCACCUCAACUUGGGUGUCGGCAUAGAGAUAUCUAUAUACAUACAUAUAUACCCAU----GUACAGCCACAGUAAUCGC-AAUAAGCCCA----------------- ..(((((......((((....(....)....)))).....((((((-((((.((-----.(((((((......)))))))((((((((....))))).....(((((........))----)))..))).)).)))))))-))).))))).----------------- ( -45.00, z-score = -3.78, R) >droEre2.scaffold_4690 7030011 112 + 18748788 UCGGGUUGAAUUUGCAUAAUUAGCAAUUAAGAUGCGCUCAAUUGCG-AUUGAUG-----AGACACCUCCGCUUGGGUGUCGGC------------------------AUAGAGCUAU----GUAUA----UAGUCAUCGC-AGUAAGCCCA----------------- ..(((((....((((.......((((((.((.....)).)))))).-...((((-----((((((((......))))))).((------------------------(((....)))----))...----...)))))))-))..))))).----------------- ( -38.60, z-score = -2.81, R) >droYak2.chrX 9502475 120 - 21770863 UCGGCUUGAAUUUGCAUAAUUAGCAAUUAAGAUGCGCUCAAUUGCG-AUUGAUG-----AGACACCUCGACUUGGGUGUCAGCAU--------------------AGAUAUACCUAU----ACACAGCCACAGUCAUGGC-AAUAAUUCCA----------------- ..((..(....((((.......((((((.((.....)).))))))(-((((.((-----.(((((((......))))))).(.((--------------------((......))))----.).....)))))))...))-))..)..)).----------------- ( -29.80, z-score = -1.09, R) >droSec1.super_56 189899 138 - 192059 UCGUGUUGAAUUUGCAUAAUUAGCAAUUAAGAUGCGCUCAAUUGCG-AUUGAUG-----AGACACCUCAACUUGGGUGUCGGCAU--AGAGAUCUAUGUACAUACAUAUAUACCCAU----GCACAGCCGCAGUCAUCGC-AAUAAUCCCA----------------- .((((((....((((.......))))....))))))....((((((-((.(((.-----.(((((((......))))))).((((--((....)))))).................(----((......)))))))))))-))).......----------------- ( -38.60, z-score = -1.92, R) >droSim1.chrX 11834235 138 - 17042790 UCGGGUUGAAUUUGCAUAAUUAGCAAUUAAGAUGCGCUCAAUUGCG-AUUGAUG-----AGGCACCUCAACUUGGGUGUCGGCAU--AGAUAUCUAUAUACAUACAUAUAUACCCAU----GCACGGCCGCAGUCAUCGC-AAUAAUCCCA----------------- ..(((......((((.......((((((.((.....)).)))))).-...((((-----((((((((......)))))))(((..--.......((((((......)))))).....----.....)))....)))))))-))....))).----------------- ( -38.01, z-score = -1.19, R) >dp4.chrXL_group1e 12245176 168 - 12523060 UGCAGGCAAAUUUGCAUAAUAAGCAAUAAGGAUGCGCUCAAUUGCACAUUCACACAUGCAACCAUGCACACUGGGGCACUGGCACGCUGGCACCCUGGCACCCUGCUACCCUGCCACCAUGGCACACUUGCACACACAUCGGACAAACACAUCGAUGUGAUGAACCCA ((((((.....((((((....(....)..(((((.((......)).)))))....))))))...(((.((.(((((((.(((((.((..(....)..))....)))))...)))).))))))))..))))))((((((((((.........)))))))).))...... ( -49.00, z-score = -0.77, R) >consensus UCGGGUUGAAUUUGCAUAAUUAGCAAUUAAGAUGCGCUCAAUUGCG_AUUGAUG_____AGACACCUCAACUUGGGUGUCGGCAU__AGA_AUCUAU_UACAUACAUAUAUACCCAU____GCACAGCCGCAGUCAUCGC_AAUAAUCCCA_________________ ............(((.......((((((.((.....)).))))))...............(((((((......))))))).))).................................................................................... (-16.76 = -16.90 + 0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:46:13 2011