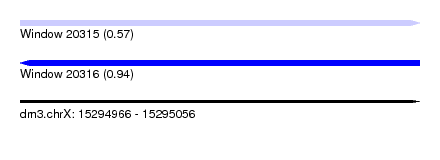
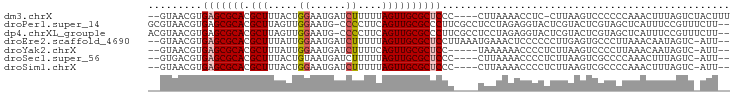
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,294,966 – 15,295,056 |
| Length | 90 |
| Max. P | 0.936772 |

| Location | 15,294,966 – 15,295,056 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 70.45 |
| Shannon entropy | 0.53978 |
| G+C content | 0.46653 |
| Mean single sequence MFE | -21.89 |
| Consensus MFE | -13.42 |
| Energy contribution | -11.75 |
| Covariance contribution | -1.67 |
| Combinations/Pair | 1.47 |
| Mean z-score | -0.70 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.568357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
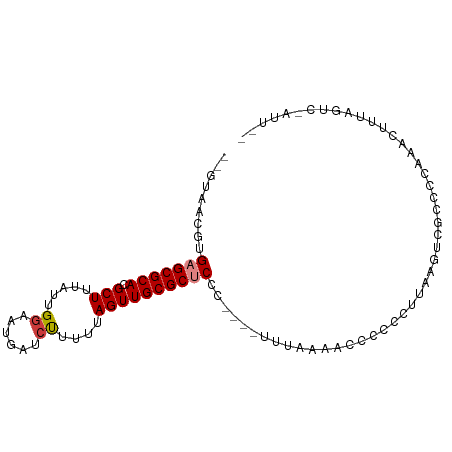
>dm3.chrX 15294966 90 + 22422827 AAAGUAGACUAAAGUUUGGGGGGACUUAAG-GAGGUUUUAAG----GGGAGCGCAACUAAAAAGAUCAUUCCAGUAAAGCGUGCGCUCACGUUAC-- .....(((((....((((((....))))))-..)))))(((.----(.(((((((........((....))..........))))))).).))).-- ( -19.47, z-score = -0.54, R) >droPer1.super_14 798577 94 - 2168203 --AAGAAACGGAAAUGAGCUACGAGUACGAGUACCUCUAGGAGGCGAAGGGCGCAACUGAAGGGG-CAUUCCAACUAAGCGUGCGCUCACGUUACGC --..(.((((....((((((((((((..((((.(((((.((..(((.....)))..))..)))))-.))))..)))...)))).))))))))).).. ( -27.30, z-score = -0.57, R) >dp4.chrXL_group1e 12235940 94 + 12523060 --AAGAAACGGAAAUGAGCUACGAGUACGAGUACCUCUAGGAGGCGAAGGGCGCAACUGAAGGGG-CAUUCCAACUAAGCGUGCGCUCACGUUACGU --..(.((((....((((((((((((..((((.(((((.((..(((.....)))..))..)))))-.))))..)))...)))).))))))))).).. ( -27.30, z-score = -0.62, R) >droEre2.scaffold_4690 7020175 92 - 18748788 --AAU-GACUAUUGUUUAAGGGCACUCAAGGGGGGAGUUUCAUUUAAGGAGCGCAACUAAAAAGAUCAUUCCAAUAAAGCGUGCGCUCACGUUAC-- --..(-(((.....((((((((.((((.......)))).)).))))))(((((((........((....))..........)))))))..)))).-- ( -18.47, z-score = -0.41, R) >droYak2.chrX 9492659 87 + 21770863 --AAU-GACUAUUGUUUAAGGGGACUUAAGAGGGGUUUUUUA-----GGAGCGCAACUGAAAAGAUCAUUCCAAUAAAGCGUGCGCUCACGUUAC-- --...-((((....((((((....))))))...)))).....-----.(((((((.((.....((....))......))..))))))).......-- ( -21.50, z-score = -1.57, R) >droSec1.super_56 179769 88 + 192059 --AAU-GACUAAAGUUUGGGGCGACUUAAGAGGGGUUUUAAG----GGGAGCGCAACUAAAAAGAUCAUUACAGUAAAGCGUGCGCUCACGUCAC-- --..(-(((.....((((((((..((....))..))))))))----..(((((((.((...................))..)))))))..)))).-- ( -19.31, z-score = -0.74, R) >droSim1.chrX 11824200 88 + 17042790 --AAU-GACUAAAGUUUGGGGCGACUUAAGAGGGGUUUUAAG----GGGAGCGCAACUAAAAAGAUCAUUCCAGUAAAGCGUGCGCUCACGUUAC-- --...-((((....((((((....))))))...)))).(((.----(.(((((((........((....))..........))))))).).))).-- ( -19.87, z-score = -0.42, R) >consensus __AAU_GACUAAAGUUUGGGGCGACUUAAGAGGGGUUUUAAA____AGGAGCGCAACUAAAAAGAUCAUUCCAAUAAAGCGUGCGCUCACGUUAC__ ......(((...............(((....)))..............(((((((.((...................))..)))))))..))).... (-13.42 = -11.75 + -1.67)
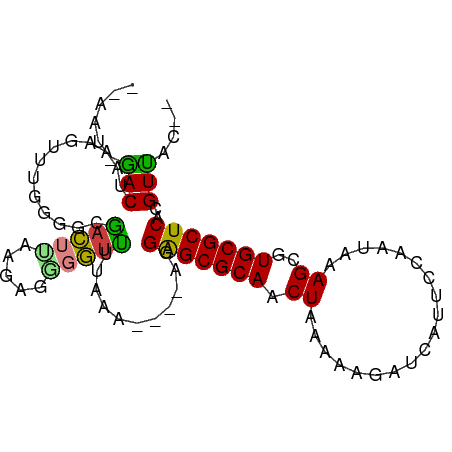
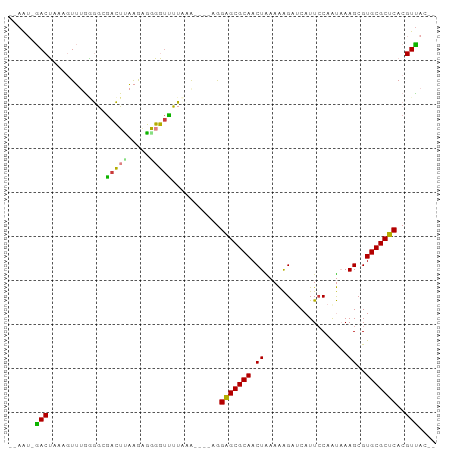
| Location | 15,294,966 – 15,295,056 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 70.45 |
| Shannon entropy | 0.53978 |
| G+C content | 0.46653 |
| Mean single sequence MFE | -18.28 |
| Consensus MFE | -11.77 |
| Energy contribution | -12.00 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.936772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15294966 90 - 22422827 --GUAACGUGAGCGCACGCUUUACUGGAAUGAUCUUUUUAGUUGCGCUCCC----CUUAAAACCUC-CUUAAGUCCCCCCAAACUUUAGUCUACUUU --(((((..(((((((......(((((((......))))))))))))))..----(((((......-.)))))...............)).)))... ( -16.90, z-score = -2.09, R) >droPer1.super_14 798577 94 + 2168203 GCGUAACGUGAGCGCACGCUUAGUUGGAAUG-CCCCUUCAGUUGCGCCCUUCGCCUCCUAGAGGUACUCGUACUCGUAGCUCAUUUCCGUUUCUU-- (((((((.(((((....)))))(..((....-..))..).))))))).............(((.(((........))).))).............-- ( -22.60, z-score = -0.05, R) >dp4.chrXL_group1e 12235940 94 - 12523060 ACGUAACGUGAGCGCACGCUUAGUUGGAAUG-CCCCUUCAGUUGCGCCCUUCGCCUCCUAGAGGUACUCGUACUCGUAGCUCAUUUCCGUUUCUU-- (((.((.((((((..(((...(((..((.((-((.((...(..(((.....)))..)..)).)))).))..)))))).)))))))).))).....-- ( -21.60, z-score = -0.01, R) >droEre2.scaffold_4690 7020175 92 + 18748788 --GUAACGUGAGCGCACGCUUUAUUGGAAUGAUCUUUUUAGUUGCGCUCCUUAAAUGAAACUCCCCCCUUGAGUGCCCUUAAACAAUAGUC-AUU-- --.....(.(((((((.(((.....(((....)))....)))))))))))...(((((.((((.......))))...............))-)))-- ( -17.19, z-score = -1.20, R) >droYak2.chrX 9492659 87 - 21770863 --GUAACGUGAGCGCACGCUUUAUUGGAAUGAUCUUUUCAGUUGCGCUCC-----UAAAAAACCCCUCUUAAGUCCCCUUAAACAAUAGUC-AUU-- --.....(.(((((((......(((((((......)))))))))))))))-----.............(((((....))))).........-...-- ( -15.90, z-score = -1.70, R) >droSec1.super_56 179769 88 - 192059 --GUGACGUGAGCGCACGCUUUACUGUAAUGAUCUUUUUAGUUGCGCUCCC----CUUAAAACCCCUCUUAAGUCGCCCCAAACUUUAGUC-AUU-- --(((((..(((((((.(((.....(.......).....))))))))))..----(((((........)))))...............)))-)).-- ( -17.80, z-score = -2.44, R) >droSim1.chrX 11824200 88 - 17042790 --GUAACGUGAGCGCACGCUUUACUGGAAUGAUCUUUUUAGUUGCGCUCCC----CUUAAAACCCCUCUUAAGUCGCCCCAAACUUUAGUC-AUU-- --.(((.(.(((((((......(((((((......)))))))))))))).)----.)))........((.((((........)))).))..-...-- ( -16.00, z-score = -1.27, R) >consensus __GUAACGUGAGCGCACGCUUUAUUGGAAUGAUCUUUUUAGUUGCGCUCCC____UUUAAAACCCCCCUUAAGUCGCCCCAAACUUUAGUC_AUU__ .........(((((((.(((.....((......))....))))))))))................................................ (-11.77 = -12.00 + 0.23)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:46:11 2011