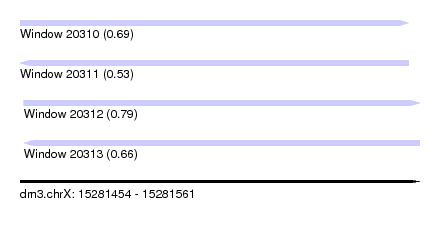
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,281,454 – 15,281,561 |
| Length | 107 |
| Max. P | 0.790408 |

| Location | 15,281,454 – 15,281,558 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 65.43 |
| Shannon entropy | 0.63872 |
| G+C content | 0.30001 |
| Mean single sequence MFE | -18.65 |
| Consensus MFE | -6.30 |
| Energy contribution | -6.92 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.693044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15281454 104 + 22422827 ------------UUUUUGAAGAAAGUCAUCGAUAAAUAUCGCUUACUUAUUAAAAUGUACAUCAAUUUGUAUGUGUUCUUACCACGAAAGGUGCGAACACUCAUUCAAGAUAUUCA ------------.(((((((((((((...((((....))))...))))........(((((......)))))((((((.((((......)))).)))))))).)))))))...... ( -24.00, z-score = -2.97, R) >droSim1.chrX 11801595 104 + 17042790 ------------UUUUUGAGGAAAGUCAUUGAUAAAUAUCGCUUACUUGUUAAAAUGUACAUAAAUUUGUAUGCGUUCGUACCACGAAAGGUGAGAACACUCAUUCAAGAUACUCA ------------....((((....(((.((((.........(((((((........(((((......)))))...((((.....))))))))))).........))))))).)))) ( -19.67, z-score = -0.65, R) >droSec1.super_56 166269 91 + 192059 ------------CUUUUGAGGAAAGCAAUCGGUAAACAUCGCUUACUUGUUAAAAUGUACAUAAAUUUGUAUGUGUUCAUA-------------AAACACUCAUUCAAGAUACUCA ------------....((((..((((.((.(.....))).)))).((((.......(((((......)))))(((((....-------------.))))).....))))...)))) ( -14.60, z-score = -0.80, R) >droYak2.chrX 9478386 104 + 21770863 ------------UUUUUGAGGAAAGUCAUCGAUAAAUAUUGCUUACUCGUUAAAAUUUGCAUAAAUUUAAAUCUGUUCCUACCACAGAAGGUAGGAACACUUUUUCAAGAUACUUG ------------.((((((((((.........(((((..(((................)))...)))))....((((((((((......)))))))))).))))))))))...... ( -24.99, z-score = -3.38, R) >droEre2.scaffold_4690 7007169 103 - 18748788 ------------UUUUUGAGGAAAGUCAGCGAUAAAUAUUGAUUACUCGUUAAAAUUUUCAUAAAUUGUAAUGCGUUCCUACCAUUUAAGGUAGAAAC-CUUUUUCAAAAUACUUG ------------.((((((((((((((((.........))))))...(((((.(((((....))))).))))).(((.(((((......))))).)))-.))))))))))...... ( -18.40, z-score = -1.61, R) >triCas2.ChLG4 6087498 108 + 13894384 UUUAAACACCAACUUUUUAAAAAAAUCAACAUUACAAACUAUAUAAUUGCAAUAAUUCACGUAAAAC-GCAAAUACUUUCAGUUCAAAAGUUGGCGA-AUUAACUCGAGA------ ........(((((((((.............................((((.................-))))..((.....))..)))))))))(((-......)))...------ ( -10.23, z-score = -0.55, R) >consensus ____________UUUUUGAGGAAAGUCAUCGAUAAAUAUCGCUUACUUGUUAAAAUGUACAUAAAUUUGUAUGUGUUCCUACCACAAAAGGUGGGAACACUCAUUCAAGAUACUCA .............(((((((..((((...((((....))))...))))........................(((((..((((......))))..)))))...)))))))...... ( -6.30 = -6.92 + 0.62)

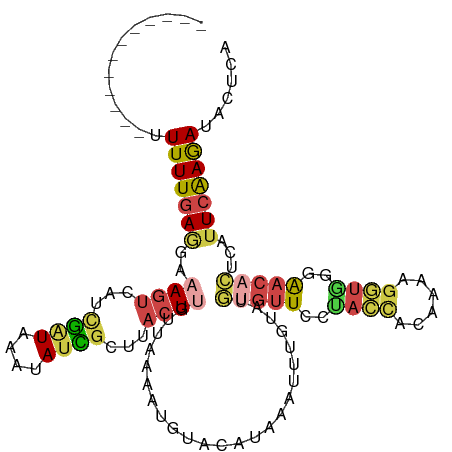
| Location | 15,281,454 – 15,281,558 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 65.43 |
| Shannon entropy | 0.63872 |
| G+C content | 0.30001 |
| Mean single sequence MFE | -21.57 |
| Consensus MFE | -5.01 |
| Energy contribution | -6.48 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.23 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.527631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15281454 104 - 22422827 UGAAUAUCUUGAAUGAGUGUUCGCACCUUUCGUGGUAAGAACACAUACAAAUUGAUGUACAUUUUAAUAAGUAAGCGAUAUUUAUCGAUGACUUUCUUCAAAAA------------ ((((......(((((.((((((..(((......)))..)))))).((((......)))))))))....((((...((((....))))...))))..))))....------------ ( -22.40, z-score = -1.83, R) >droSim1.chrX 11801595 104 - 17042790 UGAGUAUCUUGAAUGAGUGUUCUCACCUUUCGUGGUACGAACGCAUACAAAUUUAUGUACAUUUUAACAAGUAAGCGAUAUUUAUCAAUGACUUUCCUCAAAAA------------ ((((......(((((.((((((..(((......)))..)))))).((((......)))))))))....((((....(((....)))....))))..))))....------------ ( -19.10, z-score = -0.93, R) >droSec1.super_56 166269 91 - 192059 UGAGUAUCUUGAAUGAGUGUUU-------------UAUGAACACAUACAAAUUUAUGUACAUUUUAACAAGUAAGCGAUGUUUACCGAUUGCUUUCCUCAAAAG------------ ((((...((((((((.((((((-------------...)))))).((((......))))))))....)))).(((((((........)))))))..))))....------------ ( -16.30, z-score = -1.13, R) >droYak2.chrX 9478386 104 - 21770863 CAAGUAUCUUGAAAAAGUGUUCCUACCUUCUGUGGUAGGAACAGAUUUAAAUUUAUGCAAAUUUUAACGAGUAAGCAAUAUUUAUCGAUGACUUUCCUCAAAAA------------ ........((((.((((((((((((((......))))))))).((..(((((...(((..((((....))))..)))..)))))))....)))))..))))...------------ ( -23.70, z-score = -2.89, R) >droEre2.scaffold_4690 7007169 103 + 18748788 CAAGUAUUUUGAAAAAG-GUUUCUACCUUAAAUGGUAGGAACGCAUUACAAUUUAUGAAAAUUUUAACGAGUAAUCAAUAUUUAUCGCUGACUUUCCUCAAAAA------------ ......((((((.((((-(((((((((......)))))))))((.(((.(((((....))))).))).((....))..........))...))))..)))))).------------ ( -22.00, z-score = -3.07, R) >triCas2.ChLG4 6087498 108 - 13894384 ------UCUCGAGUUAAU-UCGCCAACUUUUGAACUGAAAGUAUUUGC-GUUUUACGUGAAUUAUUGCAAUUAUAUAGUUUGUAAUGUUGAUUUUUUUAAAAAGUUGGUGUUUAAA ------.......((((.-.(((((((((((.....(((((((((..(-(.....))..)))((((((((.........))))))))...))))))...))))))))))).)))). ( -25.90, z-score = -3.26, R) >consensus UGAGUAUCUUGAAUGAGUGUUCCCACCUUUCGUGGUAAGAACACAUACAAAUUUAUGUAAAUUUUAACAAGUAAGCAAUAUUUAUCGAUGACUUUCCUCAAAAA____________ ........((((....((((((..(((......)))..))))))........................((((...((((....))))...))))...))))............... ( -5.01 = -6.48 + 1.48)

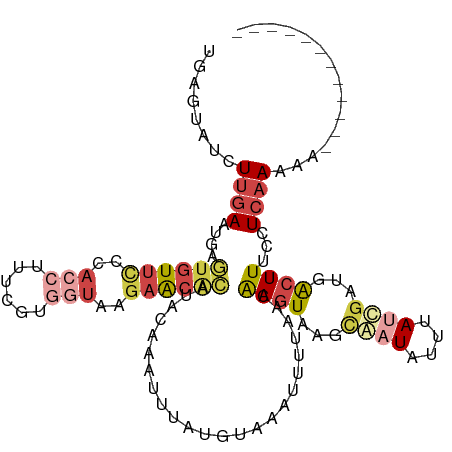
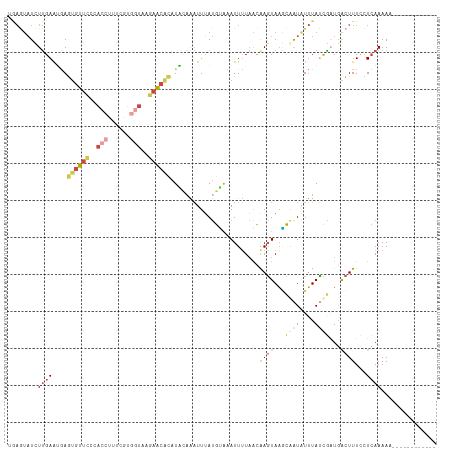
| Location | 15,281,455 – 15,281,561 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 65.79 |
| Shannon entropy | 0.64160 |
| G+C content | 0.29611 |
| Mean single sequence MFE | -19.35 |
| Consensus MFE | -6.15 |
| Energy contribution | -6.77 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.790408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
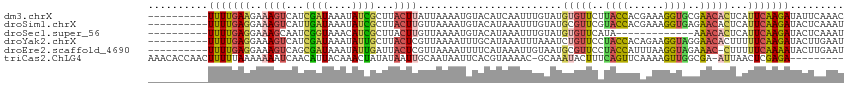
>dm3.chrX 15281455 106 + 22422827 ----------UUUUGAAGAAAGUCAUCGAUAAAUAUCGCUUACUUAUUAAAAUGUACAUCAAUUUGUAUGUGUUCUUACCACGAAAGGUGCGAACACUCAUUCAAGAUAUUCAAAC ----------(((((((((((((...((((....))))...))))........(((((......)))))((((((.((((......)))).)))))))).)))))))......... ( -23.80, z-score = -2.95, R) >droSim1.chrX 11801596 106 + 17042790 ----------UUUUGAGGAAAGUCAUUGAUAAAUAUCGCUUACUUGUUAAAAUGUACAUAAAUUUGUAUGCGUUCGUACCACGAAAGGUGAGAACACUCAUUCAAGAUACUCAAAU ----------.((((((....(((.((((.........(((((((........(((((......)))))...((((.....))))))))))).........))))))).)))))). ( -22.07, z-score = -1.60, R) >droSec1.super_56 166270 93 + 192059 ----------UUUUGAGGAAAGCAAUCGGUAAACAUCGCUUACUUGUUAAAAUGUACAUAAAUUUGUAUGUGUUCAUA-------------AAACACUCAUUCAAGAUACUCAAAU ----------.((((((..((((.((.(.....))).)))).((((.......(((((......)))))(((((....-------------.))))).....))))...)))))). ( -17.00, z-score = -1.89, R) >droYak2.chrX 9478387 106 + 21770863 ----------UUUUGAGGAAAGUCAUCGAUAAAUAUUGCUUACUCGUUAAAAUUUGCAUAAAUUUAAAUCUGUUCCUACCACAGAAGGUAGGAACACUUUUUCAAGAUACUUGAAU ----------((((((((((.........(((((..(((................)))...)))))....((((((((((......)))))))))).))))))))))......... ( -24.79, z-score = -3.01, R) >droEre2.scaffold_4690 7007170 105 - 18748788 ----------UUUUGAGGAAAGUCAGCGAUAAAUAUUGAUUACUCGUUAAAAUUUUCAUAAAUUGUAAUGCGUUCCUACCAUUUAAGGUAGAAAC-CUUUUUCAAAAUACUUGAAU ----------((((((((((((((((.........))))))...(((((.(((((....))))).))))).(((.(((((......))))).)))-.))))))))))......... ( -18.20, z-score = -1.32, R) >triCas2.ChLG4 6087501 105 + 13894384 AAACACCAACUUUUUAAAAAAAUCAACAUUACAAACUAUAUAAUUGCAAUAAUUCACGUAAAAC-GCAAAUACUUUCAGUUCAAAAGUUGGCGA-AUUAACUCGAGA--------- .....(((((((((.............................((((.................-))))..((.....))..)))))))))(((-......)))...--------- ( -10.23, z-score = -0.66, R) >consensus __________UUUUGAGGAAAGUCAUCGAUAAAUAUCGCUUACUUGUUAAAAUGUACAUAAAUUUGUAUGUGUUCCUACCACAAAAGGUGGGAACACUCAUUCAAGAUACUCAAAU ..........(((((((..((((...((((....))))...))))........................(((((..((((......))))..)))))...)))))))......... ( -6.15 = -6.77 + 0.62)
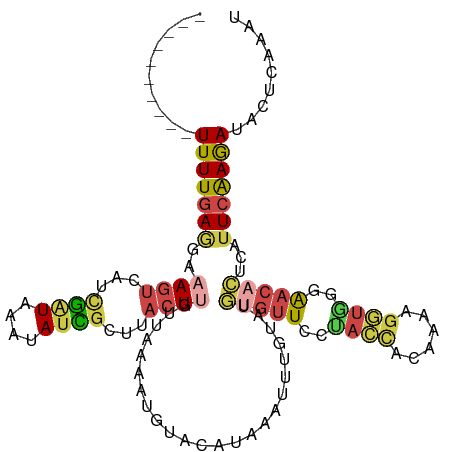

| Location | 15,281,455 – 15,281,561 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 65.79 |
| Shannon entropy | 0.64160 |
| G+C content | 0.29611 |
| Mean single sequence MFE | -22.42 |
| Consensus MFE | -5.01 |
| Energy contribution | -6.48 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.22 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.660068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
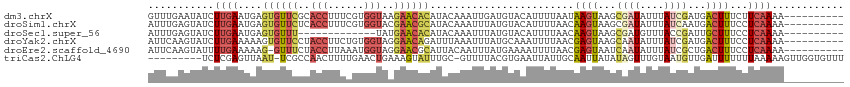
>dm3.chrX 15281455 106 - 22422827 GUUUGAAUAUCUUGAAUGAGUGUUCGCACCUUUCGUGGUAAGAACACAUACAAAUUGAUGUACAUUUUAAUAAGUAAGCGAUAUUUAUCGAUGACUUUCUUCAAAA---------- .((((((......(((((.((((((..(((......)))..)))))).((((......)))))))))....((((...((((....))))...))))..)))))).---------- ( -24.40, z-score = -2.20, R) >droSim1.chrX 11801596 106 - 17042790 AUUUGAGUAUCUUGAAUGAGUGUUCUCACCUUUCGUGGUACGAACGCAUACAAAUUUAUGUACAUUUUAACAAGUAAGCGAUAUUUAUCAAUGACUUUCCUCAAAA---------- .((((((......(((((.((((((..(((......)))..)))))).((((......)))))))))....((((....(((....)))....))))..)))))).---------- ( -21.20, z-score = -1.56, R) >droSec1.super_56 166270 93 - 192059 AUUUGAGUAUCUUGAAUGAGUGUUU-------------UAUGAACACAUACAAAUUUAUGUACAUUUUAACAAGUAAGCGAUGUUUACCGAUUGCUUUCCUCAAAA---------- .((((((...((((((((.((((((-------------...)))))).((((......))))))))....)))).(((((((........)))))))..)))))).---------- ( -18.40, z-score = -1.89, R) >droYak2.chrX 9478387 106 - 21770863 AUUCAAGUAUCUUGAAAAAGUGUUCCUACCUUCUGUGGUAGGAACAGAUUUAAAUUUAUGCAAAUUUUAACGAGUAAGCAAUAUUUAUCGAUGACUUUCCUCAAAA---------- .((((((...))))))((((((((((((((......))))))))).((..(((((...(((..((((....))))..)))..)))))))....)))))........---------- ( -24.30, z-score = -2.91, R) >droEre2.scaffold_4690 7007170 105 + 18748788 AUUCAAGUAUUUUGAAAAAG-GUUUCUACCUUAAAUGGUAGGAACGCAUUACAAUUUAUGAAAAUUUUAACGAGUAAUCAAUAUUUAUCGCUGACUUUCCUCAAAA---------- .........((((((.((((-(((((((((......)))))))))((.(((.(((((....))))).))).((....))..........))...))))..))))))---------- ( -21.20, z-score = -2.65, R) >triCas2.ChLG4 6087501 105 - 13894384 ---------UCUCGAGUUAAU-UCGCCAACUUUUGAACUGAAAGUAUUUGC-GUUUUACGUGAAUUAUUGCAAUUAUAUAGUUUGUAAUGUUGAUUUUUUUAAAAAGUUGGUGUUU ---------............-.(((((((((((.....(((((((((..(-(.....))..)))((((((((.........))))))))...))))))...)))))))))))... ( -25.00, z-score = -3.09, R) >consensus AUUUGAGUAUCUUGAAUGAGUGUUCCCACCUUUCGUGGUAAGAACACAUACAAAUUUAUGUAAAUUUUAACAAGUAAGCAAUAUUUAUCGAUGACUUUCCUCAAAA__________ ...........((((....((((((..(((......)))..))))))........................((((...((((....))))...))))...))))............ ( -5.01 = -6.48 + 1.48)
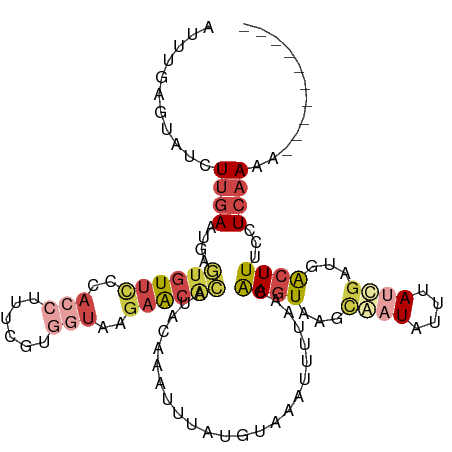
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:46:08 2011