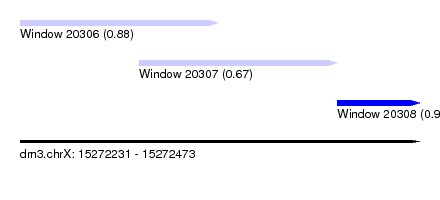
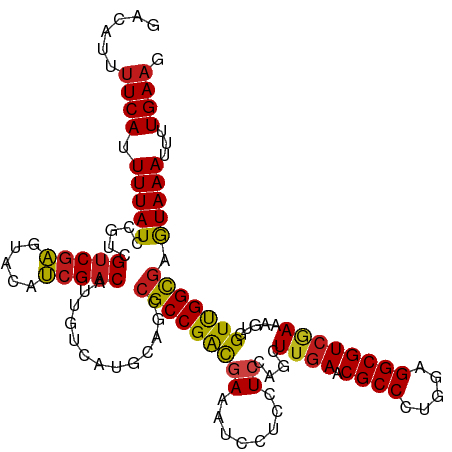
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,272,231 – 15,272,473 |
| Length | 242 |
| Max. P | 0.968419 |

| Location | 15,272,231 – 15,272,351 |
|---|---|
| Length | 120 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.22 |
| Shannon entropy | 0.18466 |
| G+C content | 0.50462 |
| Mean single sequence MFE | -41.45 |
| Consensus MFE | -29.11 |
| Energy contribution | -29.85 |
| Covariance contribution | 0.74 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.881488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
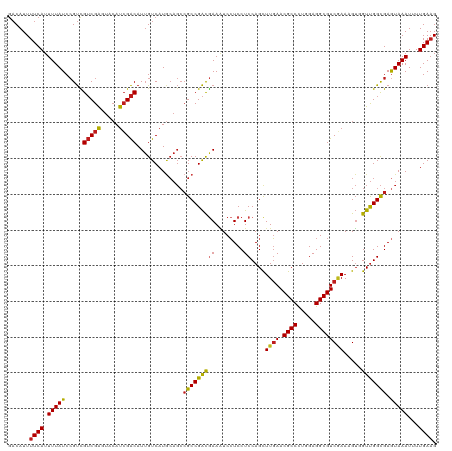
>dm3.chrX 15272231 120 + 22422827 GGCAUGCAGCGACGCAUUUGCUCCGAACUUCGAUAGGAGGGACUGUAGCGUCCUGGAAAACAACUGGGCGCUGACAUUUUCAUUUUAUCCGUCGUCGAAUACAUCGACAUUGUCAUGCAG .((((((((.(((((....))..........((((((((..(.((((((((((.(........).))))))).))).)..).))))))).)))(((((.....))))).))).))))).. ( -41.40, z-score = -2.75, R) >droWil1.scaffold_180702 2791570 120 - 4511350 GGCAUACAGCGUCGCAUUUGUUCUGAACUUCGAUACGAUGGACUGUAGCGUCCUGGAAAACAACUGGGCGCUGACAUUUUCAUUUUAUCCGUUGUCGAAUACAUCGACAUUGUCAUGCAG .((((((((.((((.((.(((((.((.....((((.((((((.((((((((((.(........).))))))).)))..)))))).)))).....))))))).)))))).)))).)))).. ( -39.30, z-score = -2.96, R) >droSim1.chrX 11791864 120 + 17042790 GGCAUGCAGCGACGCAUUUGCUCCGAACUUCGAUAGGAGGGACUGUAGCGUCCUGGAAAACAACUGGGCGCUGACAUUUUCAUUUUAUCCGUCGUCGAGUACAUCGACAUUGUCAUGCAG .((((((((.(((((....))..........((((((((..(.((((((((((.(........).))))))).))).)..).))))))).)))(((((.....))))).))).))))).. ( -42.20, z-score = -2.64, R) >droSec1.super_56 157159 120 + 192059 GGCAUGCAGCGACGCAUUUGCUCCGAACUUCGAUAGGAGGGACUGUAGCGUCCUGGAAAACAACUGGGCGCUGACAUUUUCAUUUUAUCCGUCGUCGAGUACAUCGACAUUGUCAUGCAG .((((((((.(((((....))..........((((((((..(.((((((((((.(........).))))))).))).)..).))))))).)))(((((.....))))).))).))))).. ( -42.20, z-score = -2.64, R) >droYak2.chrX 9468759 120 + 21770863 GGCAUGCAGCGACGCAUUUGCUCCGAACUUCGAUAGGAGGGACUGUAGCGUCCUGGAAAACAACUGGGCGCUGACAUUUUCAUUUUAUCCGUCGUCGAGUACAUCGACAUUGUCAUGCAG .((((((((.(((((....))..........((((((((..(.((((((((((.(........).))))))).))).)..).))))))).)))(((((.....))))).))).))))).. ( -42.20, z-score = -2.64, R) >droEre2.scaffold_4690 6997945 120 - 18748788 GGCAUGCAGCGACGCAUUUGCUCCGAACUUCGAUAGGAGGGACUGUAGCGUCCUGGAAAACAACUGGGCGCUGACAUUUUCAUUUUAUCCGUCGUCGAGUACAUCGACAUUGUCAUGCAG .((((((((.(((((....))..........((((((((..(.((((((((((.(........).))))))).))).)..).))))))).)))(((((.....))))).))).))))).. ( -42.20, z-score = -2.64, R) >droAna3.scaffold_13117 1488283 120 + 5790199 GGCAUGCAGCGGCGCAUCUGCUCCGAACUCCGAUAGGAGGGACUGUAGCGUCCAGGAAAACAACUGGGCGCUGACAUUUUCAUUUUAUCCGUCGUCGAGUACAUCGACAUUGUCAUGCAG .(((((((((((.((....)).)))......((((((((..(.((((((((((((........))))))))).))).)..).)))))))....(((((.....))))).))).))))).. ( -47.90, z-score = -3.75, R) >dp4.chrXL_group1e 12212544 120 + 12523060 GGCAUGCAGCGCCGCAUUUGCUCCGAACUUCGAUAGGAGGGACUGUAGCGUCCUGGAAAACAACUGGGCGCUGACAUUUUCAUUUUAUCCGUUGUCGAGUACAUCGACAUUGUCAUGCAG .((((((((((((.((.(((.((((..((((....))))((((......))))))))...))).)))))))))................((.((((((.....)))))).)).))))).. ( -42.70, z-score = -2.64, R) >droPer1.super_14 774364 120 - 2168203 GGCAUGCAGCGCCGCAUUUGCUCCGAACUUCGAUAGGAGGGACUGUAGCGUCCUGGAAAACAACUGGGCGCUGACAUUUUCAUUUUAUCCGUUGUCGAGUACAUCGACAUUGUCAUGCAG .((((((((((((.((.(((.((((..((((....))))((((......))))))))...))).)))))))))................((.((((((.....)))))).)).))))).. ( -42.70, z-score = -2.64, R) >droVir3.scaffold_12928 7300914 120 + 7717345 GGCAUACAGCGCCGCAUUUGCUCCGAACUUCGAUAGGAUGGACUGUAGCGUCCUGGAAAACAACUGGGCGCUGACAUUUUCAUUUUAUCCGUCGUCGAAUACAUCGACAUUGUCAUGCAG .((((((((((..((....))..))......(((((((((((.((((((((((.(........).))))))).)))..)))))))))))....(((((.....))))).)))).)))).. ( -38.20, z-score = -2.24, R) >droGri2.scaffold_15081 864217 120 - 4274704 GGCAUACAGCGUCGCAUCUGCUCCGAACUUCGAUAGGAUGGACUGUAGCGUCCUGGAAAACAACUGGGCGCUGACAUUUUCAUUUUAUCCGUUGUCGAAUACAUCGACAUUGUCAUACAG ((((.....((..((....))..))......(((((((((((.((((((((((.(........).))))))).)))..)))))))))))...((((((.....)))))).))))...... ( -36.70, z-score = -2.32, R) >droMoj3.scaffold_6473 14179207 120 - 16943266 GGCAUGCAGCGUCGCAUUUGCUCCGAACUUCGAUAGGAUGGACUGUAGCGUCCUGGAAAACAACUGGGCGCUGACAUUUUCAUUUUAUCCGUCGUCGAGUACAUCGACAUUGUCAUGCAG .((((((((.((((.((.(((((.((.....(((((((((((.((((((((((.(........).))))))).)))..))))))))))).....))))))).)))))).))).))))).. ( -46.40, z-score = -3.89, R) >anoGam1.chr2R 45868014 116 - 62725911 GCCAUACACCGUCGCAUCUGCUCGGGCCCCCUGUAGGUGGGGCUGUAUCGACCGGGAAAGCAGCUCGGAGUACUCAUUUUCAUUUUAUCGGCUGACGGAUAAACGGGCAUAGUCAU---- ((((((((..(((.(((((((..((....)).))))))).))))))))((.(((((.......)))))...............((((((.(....).)))))))))))........---- ( -34.80, z-score = 0.50, R) >consensus GGCAUGCAGCGACGCAUUUGCUCCGAACUUCGAUAGGAGGGACUGUAGCGUCCUGGAAAACAACUGGGCGCUGACAUUUUCAUUUUAUCCGUCGUCGAGUACAUCGACAUUGUCAUGCAG .((((((((((..((....))..........(((((((.(((.((((((((((.(........).))))))).)))..))).)))))))))).(((((.....)))))..))).)))).. (-29.11 = -29.85 + 0.74)

| Location | 15,272,303 – 15,272,423 |
|---|---|
| Length | 120 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.84 |
| Shannon entropy | 0.16588 |
| G+C content | 0.48519 |
| Mean single sequence MFE | -36.01 |
| Consensus MFE | -26.42 |
| Energy contribution | -26.42 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.667957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
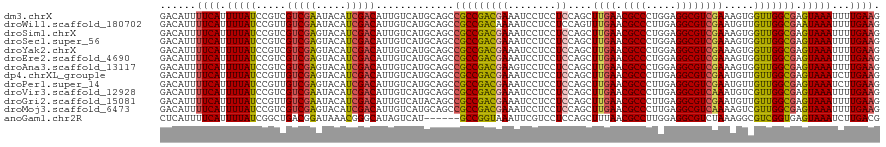
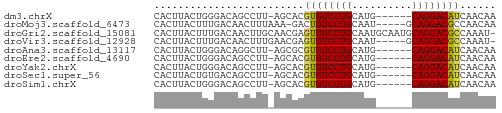
>dm3.chrX 15272303 120 + 22422827 GACAUUUUCAUUUUAUCCGUCGUCGAAUACAUCGACAUUGUCAUGCAGCCGCCGACGAAAUCCUCCUCCAGCUUGAACGCCCUGGAGGCGUCGAAAGUGGUUGGCGAGUAAAUUUUGAAG ......((((.((((..(((((((((.....)))))..........(((((((((((.......(((((((..........))))))))))))...))))))))))..))))...)))). ( -38.41, z-score = -2.47, R) >droWil1.scaffold_180702 2791642 120 - 4511350 GACAUUUUCAUUUUAUCCGUUGUCGAAUACAUCGACAUUGUCAUGCAGCCGCCGACAAAAUCCUCCUCCAGUUUGAACGCCCUUGAGGCGUCGAAUGUUGUUGGCGAAUAAAUUUUGAAG ......((((.(((((.((.((((((.....)))))).)).........(((((((((............((((((.((((.....)))))))))).))))))))).)))))...)))). ( -33.22, z-score = -2.47, R) >droSim1.chrX 11791936 120 + 17042790 GACAUUUUCAUUUUAUCCGUCGUCGAGUACAUCGACAUUGUCAUGCAGCCGCCGACGAAAUCCUCCUCCAGCUUGAACGCCCUGGAGGCGUCGAAAGUGGUUGGCGAGUAAAUUUUGAAG ......((((.((((..(((((((((.....)))))..........(((((((((((.......(((((((..........))))))))))))...))))))))))..))))...)))). ( -39.21, z-score = -2.48, R) >droSec1.super_56 157231 120 + 192059 GACAUUUUCAUUUUAUCCGUCGUCGAGUACAUCGACAUUGUCAUGCAGCCGCCGACGAAAUCCUCCUCCAGCUUGAACGCCCUGGAGGCGUCGAAAGUGGUUGGCGAGUAAAUUUUGAAG ......((((.((((..(((((((((.....)))))..........(((((((((((.......(((((((..........))))))))))))...))))))))))..))))...)))). ( -39.21, z-score = -2.48, R) >droYak2.chrX 9468831 120 + 21770863 GACAUUUUCAUUUUAUCCGUCGUCGAGUACAUCGACAUUGUCAUGCAGCCGCCGACGAAAUCCUCCUCCAGCUUGAACGCCCUGGAGGCGUCGAAAGUGGUUGGCGAGUAAAUUUUGAAG ......((((.((((..(((((((((.....)))))..........(((((((((((.......(((((((..........))))))))))))...))))))))))..))))...)))). ( -39.21, z-score = -2.48, R) >droEre2.scaffold_4690 6998017 120 - 18748788 GACAUUUUCAUUUUAUCCGUCGUCGAGUACAUCGACAUUGUCAUGCAGCCGCCGACGAAAUCCUCCUCCAGCUUGAACGCCCUGGAGGCGUCGAAAGUGGUUGGCGAGUAAAUUUUGAAG ......((((.((((..(((((((((.....)))))..........(((((((((((.......(((((((..........))))))))))))...))))))))))..))))...)))). ( -39.21, z-score = -2.48, R) >droAna3.scaffold_13117 1488355 120 + 5790199 GACAUUUUCAUUUUAUCCGUCGUCGAGUACAUCGACAUUGUCAUGCAGCCGCCGACGAAGUCCUCCUCCAGCUUGAACGCCCUGGAGGCGUCGAAAGUGGUUGGCGAGUAAAUUUUGAAG ......((((.((((..(((((((((.....)))))..........(((((((((((.......(((((((..........))))))))))))...))))))))))..))))...)))). ( -39.21, z-score = -2.25, R) >dp4.chrXL_group1e 12212616 120 + 12523060 GACAUUUUCAUUUUAUCCGUUGUCGAGUACAUCGACAUUGUCAUGCAGCCGCCGACGAAAUCCUCCUCCAGCUUGAACGCCCUUGAGGCGUCGAAUGUUGUUGGCGAGUAAAUCUUGAAG ......((((.(((((....((((((.....))))))((((((.(((((...(((((....((((.....((......))....)))))))))...))))))))))))))))...)))). ( -32.30, z-score = -1.24, R) >droPer1.super_14 774436 120 - 2168203 GACAUUUUCAUUUUAUCCGUUGUCGAGUACAUCGACAUUGUCAUGCAGCCGCCGACGAAAUCCUCCUCCAGCUUGAACGCCCUUGAGGCGUCGAAUGUUGUUGGCGAGUAAAUCUUGAAG ......((((.(((((....((((((.....))))))((((((.(((((...(((((....((((.....((......))....)))))))))...))))))))))))))))...)))). ( -32.30, z-score = -1.24, R) >droVir3.scaffold_12928 7300986 120 + 7717345 GACAUUUUCAUUUUAUCCGUCGUCGAAUACAUCGACAUUGUCAUGCAGCCGCCGACGAAAUCCUCCUCCAGCUUGAACGCCCUUGAGGCGUCAAAUGUCGUUGGCGAGUAAAUUUUGAAG ......((((.(((((.....(((((.....))))).............(((((((((..........((..((((.((((.....)))))))).))))))))))).)))))...)))). ( -32.20, z-score = -1.65, R) >droGri2.scaffold_15081 864289 120 - 4274704 GACAUUUUCAUUUUAUCCGUUGUCGAAUACAUCGACAUUGUCAUACAGCCGCCGACGAAAUCCUCCUCCAGCUUGAACGCCCUUGAGGCGUCGAAUGUUGUUGGCGAGUAAAUUUUGAAG ......((((.(((((....((((((.....))))))((((((.(((((...(((((....((((.....((......))....)))))))))...))))))))))))))))...)))). ( -31.50, z-score = -1.76, R) >droMoj3.scaffold_6473 14179279 120 - 16943266 GACAUUUUCAUUUUAUCCGUCGUCGAGUACAUCGACAUUGUCAUGCAGCCGCCGACGAAAUCCUCCUCCAGCUUGAACGCCCUUGAGGCGUCAAAAGUCGUUGGCGAGUAAAUUUUGAAG ......((((.(((((.....(((((.....))))).............(((((((((..............((((.((((.....))))))))...))))))))).)))))...)))). ( -32.03, z-score = -1.47, R) >anoGam1.chr2R 45868086 114 - 62725911 CUCAUUUUCAUUUUAUCGGCUGACGGAUAAACGGGCAUAGUCAU------GCCGGUAAAUUCGUCCUCCAGCUUUAACGCCUUGGAGGCGUCUAAAGGCGUCGGUGAGUAAAUCUUGACG .(((.......(((((((((.(((((((..((.(((((....))------))).))..))))))).....(((((.(((((.....)))))...)))))))))))))).......))).. ( -40.14, z-score = -3.44, R) >consensus GACAUUUUCAUUUUAUCCGUCGUCGAGUACAUCGACAUUGUCAUGCAGCCGCCGACGAAAUCCUCCUCCAGCUUGAACGCCCUGGAGGCGUCGAAAGUGGUUGGCGAGUAAAUUUUGAAG ......((((.(((((.....(((((.....))))).............(((((((((........))....((((.((((.....)))))))).....))))))).)))))...)))). (-26.42 = -26.42 + 0.00)
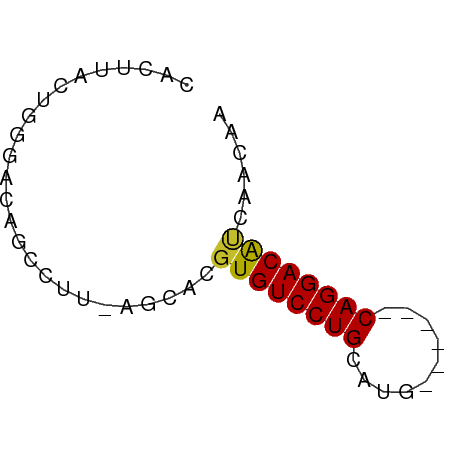
| Location | 15,272,423 – 15,272,473 |
|---|---|
| Length | 50 |
| Sequences | 9 |
| Columns | 57 |
| Reading direction | forward |
| Mean pairwise identity | 81.13 |
| Shannon entropy | 0.34094 |
| G+C content | 0.49629 |
| Mean single sequence MFE | -15.07 |
| Consensus MFE | -10.58 |
| Energy contribution | -10.32 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.968419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15272423 50 + 22422827 CACUUACUGGGACAGCCUU-AGCACGUGUCCUGCAUG------CAGGACAUCAACAA ......(((((.....)))-))...((((((((....------))))))))...... ( -14.80, z-score = -1.70, R) >droMoj3.scaffold_6473 14179399 51 - 16943266 CACUUACUUUGACAACUUUAAA-GACUGUCCUGCAAU-----GCAGGACGCCAACAA ......((((((.....)))))-)...(((((((...-----)))))))........ ( -13.90, z-score = -3.94, R) >droGri2.scaffold_15081 864409 56 - 4274704 CACUUACUUUGACAACUUGCAACGAGUGUCCUGCAAUGCAAUGCAGGACGCCAAAU- .......................(.((((((((((......)))))))))))....- ( -17.90, z-score = -3.34, R) >droVir3.scaffold_12928 7301106 51 + 7717345 CACUUACUUUGACAACUUUGAACGAGUGUCCUGCAAU-----GCAGGACGCCAAAU- .......................(.(((((((((...-----))))))))))....- ( -15.20, z-score = -3.53, R) >droAna3.scaffold_13117 1488475 50 + 5790199 CACUUACUGGGACAGGCUU-AGCGCGUGUCCUGCAUG------CAGGACAUCAACAA ......(((((.....)))-))...((((((((....------))))))))...... ( -14.60, z-score = -1.17, R) >droEre2.scaffold_4690 6998137 50 - 18748788 CACUUACUGGGACAGCCUU-AGCACGUGUCCUGCAUG------CAGGACAUCAACAA ......(((((.....)))-))...((((((((....------))))))))...... ( -14.80, z-score = -1.70, R) >droYak2.chrX 9468951 50 + 21770863 CACUUACUGGGACAGCCUU-AGCACGUGUCCUGCAUG------CAGGACAUCAACAA ......(((((.....)))-))...((((((((....------))))))))...... ( -14.80, z-score = -1.70, R) >droSec1.super_56 157351 50 + 192059 CACUUACUGUGACAGCCUU-AGCACGUGUCCUGCAUG------CAGGACAUCAACAA (((.....)))...((...-.))..((((((((....------))))))))...... ( -14.80, z-score = -1.74, R) >droSim1.chrX 11792056 50 + 17042790 CACUUACUGGGACAGCCUU-AGCACGUGUCCUGCAUG------CAGGACAUCAACAA ......(((((.....)))-))...((((((((....------))))))))...... ( -14.80, z-score = -1.70, R) >consensus CACUUACUGGGACAGCCUU_AGCACGUGUCCUGCAUG______CAGGACAUCAACAA .........................((((((((..........))))))))...... (-10.58 = -10.32 + -0.26)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:46:05 2011