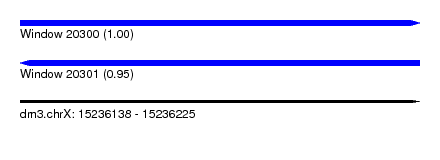
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,236,138 – 15,236,225 |
| Length | 87 |
| Max. P | 0.999914 |

| Location | 15,236,138 – 15,236,225 |
|---|---|
| Length | 87 |
| Sequences | 5 |
| Columns | 89 |
| Reading direction | forward |
| Mean pairwise identity | 65.00 |
| Shannon entropy | 0.63266 |
| G+C content | 0.38014 |
| Mean single sequence MFE | -27.72 |
| Consensus MFE | -11.74 |
| Energy contribution | -12.42 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.45 |
| Mean z-score | -4.10 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.86 |
| SVM RNA-class probability | 0.999914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
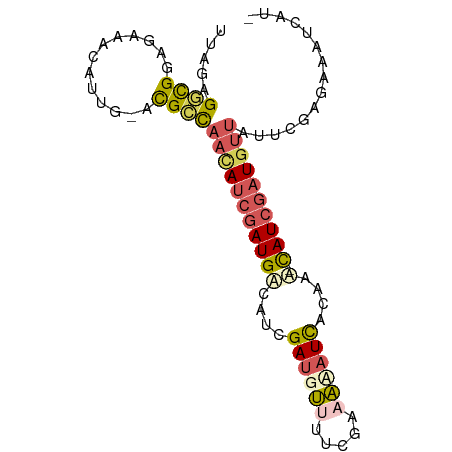
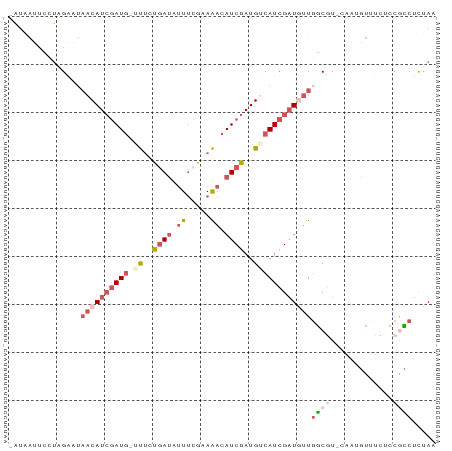
>dm3.chrX 15236138 87 + 22422827 UUAGAGGCGGAGAAACUUUUUACGCCAACAUCGAUGACAUCGAUGUUUUCGAAAUAUCAGAAA-CAUCGAUGUUUUUCUAGGAAUGUU- (((((((((.............)))).........(((((((((((((..((....))..)))-))))))))))..))))).......- ( -27.62, z-score = -4.11, R) >droYak2.chr3L 279695 87 - 24197627 UUAGUGAUGGGCAUAUGACG-AUGCCAAAAACAAUGUCAUCGAUGUUUUGUAAACAUCGGCAA-UAUAUAUGUGAAGCUCAAUAUUAAA ((((((.(((((..((((((-.((.......)).))))))(((((((.....)))))))....-............))))).)))))). ( -22.00, z-score = -2.19, R) >droSec1.super_56 123725 87 + 192059 CUAAAGGCGGAGAAACAUUG-ACGCCAACAUCGAUGACAUCGAUGUCUACGAAAAAUCACAGU-CAUCGAUGUUAUUCGAGAAAUCAUC .....((((..((....)).-.))))....(((((((((((((((.((..((....))..)).-))))))))))).))))......... ( -28.60, z-score = -4.23, R) >droSim1.chrX 11759212 87 + 17042790 CUAAAGGCGGAGAAACAUUG-ACGCCAACAUCGAUGACAUCGAUGUCUACGAAAAAUCACAGU-CAUCGAUGUUAUUCGAGGAAUCAUC .....((((..((....)).-.))))....(((((((((((((((.((..((....))..)).-))))))))))).))))......... ( -28.60, z-score = -3.94, R) >droWil1.scaffold_181108 3252493 86 - 4707319 UUAGAGCUGGAAAAAACAACGAUGGUACUAUCGAUG-CAUCGAUGGUUUUGUAUUUUUAUAAAACAUCGAUGUUUGGCCAGCACAUA-- .....(((((...(((((.(((((..((((((((..-..))))))))((((((....)))))).))))).)))))..))))).....-- ( -31.80, z-score = -6.03, R) >consensus UUAGAGGCGGAGAAACAUUG_ACGCCAACAUCGAUGACAUCGAUGUUUUCGAAAAAUCACAAA_CAUCGAUGUUAUUCGAGAAAUCAU_ .....((((.............))))((((((((((.....((((((.....))))))......))))))))))............... (-11.74 = -12.42 + 0.68)

| Location | 15,236,138 – 15,236,225 |
|---|---|
| Length | 87 |
| Sequences | 5 |
| Columns | 89 |
| Reading direction | reverse |
| Mean pairwise identity | 65.00 |
| Shannon entropy | 0.63266 |
| G+C content | 0.38014 |
| Mean single sequence MFE | -22.12 |
| Consensus MFE | -6.60 |
| Energy contribution | -6.48 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.952958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
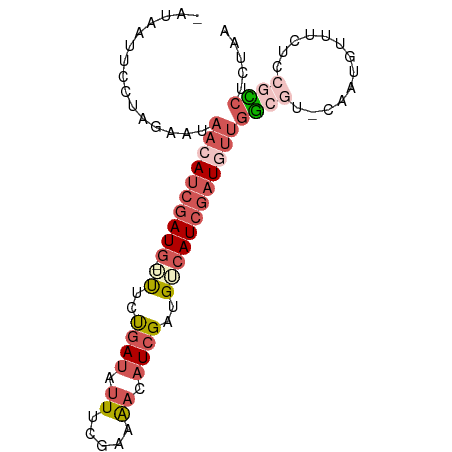
>dm3.chrX 15236138 87 - 22422827 -AACAUUCCUAGAAAAACAUCGAUG-UUUCUGAUAUUUCGAAAACAUCGAUGUCAUCGAUGUUGGCGUAAAAAGUUUCUCCGCCUCUAA -((((((....((...(((((((((-(((.(((....))).))))))))))))..))))))))((((.............))))..... ( -24.32, z-score = -4.19, R) >droYak2.chr3L 279695 87 + 24197627 UUUAAUAUUGAGCUUCACAUAUAUA-UUGCCGAUGUUUACAAAACAUCGAUGACAUUGUUUUUGGCAU-CGUCAUAUGCCCAUCACUAA ........((.((............-....(((((((.....)))))))(((((..(((.....))).-.)))))..)).))....... ( -16.00, z-score = -1.25, R) >droSec1.super_56 123725 87 - 192059 GAUGAUUUCUCGAAUAACAUCGAUG-ACUGUGAUUUUUCGUAGACAUCGAUGUCAUCGAUGUUGGCGU-CAAUGUUUCUCCGCCUUUAG .........((((...(((((((((-.((((((....)))))).)))))))))..))))((..((((.-...........))))..)). ( -24.70, z-score = -1.86, R) >droSim1.chrX 11759212 87 - 17042790 GAUGAUUCCUCGAAUAACAUCGAUG-ACUGUGAUUUUUCGUAGACAUCGAUGUCAUCGAUGUUGGCGU-CAAUGUUUCUCCGCCUUUAG .........((((...(((((((((-.((((((....)))))).)))))))))..))))((..((((.-...........))))..)). ( -24.70, z-score = -1.93, R) >droWil1.scaffold_181108 3252493 86 + 4707319 --UAUGUGCUGGCCAAACAUCGAUGUUUUAUAAAAAUACAAAACCAUCGAUG-CAUCGAUAGUACCAUCGUUGUUUUUUCCAGCUCUAA --...(.(((((..(((((.(((((...(((....)))....((.(((((..-..))))).))..))))).)))))...))))).)... ( -20.90, z-score = -3.37, R) >consensus _AUAAUUCCUAGAAUAACAUCGAUG_UUUCUGAUAUUUCGAAAACAUCGAUGUCAUCGAUGUUGGCGU_CAAUGUUUCUCCGCCUCUAA ..........................................(((((((((...))))))))).......................... ( -6.60 = -6.48 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:45:59 2011