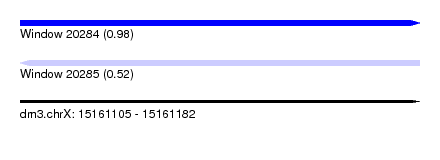
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,161,105 – 15,161,182 |
| Length | 77 |
| Max. P | 0.977590 |

| Location | 15,161,105 – 15,161,182 |
|---|---|
| Length | 77 |
| Sequences | 10 |
| Columns | 81 |
| Reading direction | forward |
| Mean pairwise identity | 70.42 |
| Shannon entropy | 0.58451 |
| G+C content | 0.49094 |
| Mean single sequence MFE | -14.09 |
| Consensus MFE | -6.94 |
| Energy contribution | -7.14 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.977590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
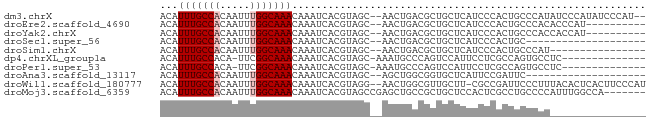
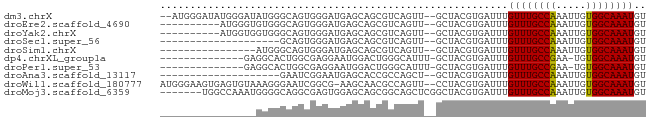
>dm3.chrX 15161105 77 + 22422827 ACAUUUGCCACAAUUUGGCAAACAAAUCACGUAGC--AACUGACGCUGCUCAUCCCACUGCCCAUAUCCCAUAUCCCAU-- ...(((((((.....)))))))........(((((--.......)))))..............................-- ( -13.70, z-score = -2.48, R) >droEre2.scaffold_4690 6891689 69 - 18748788 ACAUUUGCCACAAUUUGGCAAACAAAUCACGUAGC--AACUGACGCUGCUCAUCCCACUGCCCACACCCAU---------- ...(((((((.....)))))))........(((((--.......)))))......................---------- ( -13.70, z-score = -2.35, R) >droYak2.chrX 9361137 69 + 21770863 ACAUUUGCCACAAUUUGGCAAACAAAUCACGUAGC--AACUGACGCUGCUCAUCCCACUGCCCACCACCAU---------- ...(((((((.....)))))))........(((((--.......)))))......................---------- ( -13.70, z-score = -2.35, R) >droSec1.super_56 50713 59 + 192059 ACAUUUGCCACAAUUUGGCAAACAAAUCACGUAGC--AACUGACGCUGCUCAUCCCACUGC-------------------- ...(((((((.....)))))))........(((((--.......)))))............-------------------- ( -13.70, z-score = -2.33, R) >droSim1.chrX 11698710 63 + 17042790 ACAUUUGCCACAAUUUGGCAAACAAAUCACGUAGC--AACUGACGCUGCUCAUCCCACUGCCCAU---------------- ...(((((((.....)))))))........(((((--.......)))))................---------------- ( -13.70, z-score = -2.26, R) >dp4.chrXL_group1a 7508126 65 - 9151740 ACAUUUGCCACA-UUCGGCAAACAAAUCACGUAGC-AAAUGCCCAGUCCAUUCCUCGCCAGUGCCUC-------------- ...((((((...-...))))))...........((-(...((...(......)...))...)))...-------------- ( -9.40, z-score = -0.61, R) >droPer1.super_53 110941 65 + 525408 ACAUUUGCCACA-UUCGGCAAACAAAUCACGUAGC-AAAUGCCCAGUCCAUUCCUCGCCAGUGCCUC-------------- ...((((((...-...))))))...........((-(...((...(......)...))...)))...-------------- ( -9.40, z-score = -0.61, R) >droAna3.scaffold_13117 4411075 59 - 5790199 ACAUUUGCCACAAUUUGGCAAACAAAUCACGUAGC--AGCUGGCGGUGCUCAUUCCGAUUC-------------------- ...(((((((.....)))))))........(.(((--(.(....).)))))..........-------------------- ( -12.60, z-score = -0.52, R) >droWil1.scaffold_180777 2241000 78 + 4753960 ACAUUUGCCACAAUUUGGCAAACAAAUCACGUAGG--AACUGGCGUUGCUU-CGCCGAUUCCCUUUACACUCACUUCCCAU ...(((((((.....)))))))........(((((--((.(((((......-))))).))))...)))............. ( -20.90, z-score = -3.85, R) >droMoj3.scaffold_6359 2010207 74 + 4525533 ACAUUUGCCACAAUUUGGCAAACAAAUCACGUAGCCGAGCUGCCGCUGCUCCACUCGCCUGCCCCAUUUGGCCA------- ...(((((((.....)))))))(((((...((((.((((..((....))....)))).))))...)))))....------- ( -20.10, z-score = -1.53, R) >consensus ACAUUUGCCACAAUUUGGCAAACAAAUCACGUAGC__AACUGACGCUGCUCAUCCCACUGCCCAC________________ ...(((((((.....)))))))........................................................... ( -6.94 = -7.14 + 0.20)
| Location | 15,161,105 – 15,161,182 |
|---|---|
| Length | 77 |
| Sequences | 10 |
| Columns | 81 |
| Reading direction | reverse |
| Mean pairwise identity | 70.42 |
| Shannon entropy | 0.58451 |
| G+C content | 0.49094 |
| Mean single sequence MFE | -19.73 |
| Consensus MFE | -9.22 |
| Energy contribution | -9.06 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.519992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
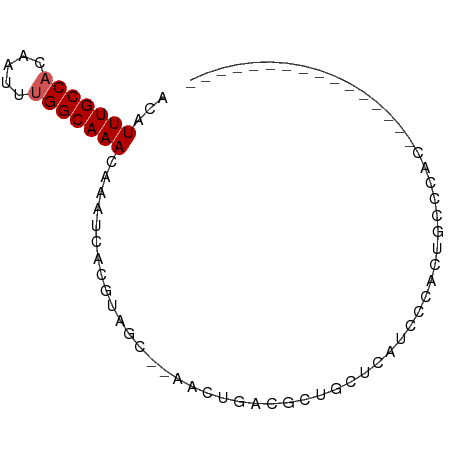
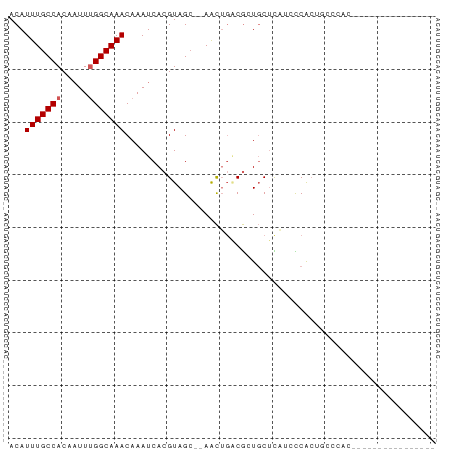
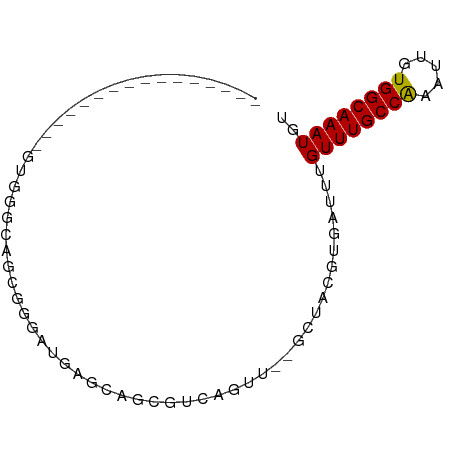
>dm3.chrX 15161105 77 - 22422827 --AUGGGAUAUGGGAUAUGGGCAGUGGGAUGAGCAGCGUCAGUU--GCUACGUGAUUUGUUUGCCAAAUUGUGGCAAAUGU --..........................(((((((((....)))--))).))).....((((((((.....)))))))).. ( -18.50, z-score = -0.13, R) >droEre2.scaffold_4690 6891689 69 + 18748788 ----------AUGGGUGUGGGCAGUGGGAUGAGCAGCGUCAGUU--GCUACGUGAUUUGUUUGCCAAAUUGUGGCAAAUGU ----------.(.(.((....)).).).(((((((((....)))--))).))).....((((((((.....)))))))).. ( -18.60, z-score = -0.29, R) >droYak2.chrX 9361137 69 - 21770863 ----------AUGGUGGUGGGCAGUGGGAUGAGCAGCGUCAGUU--GCUACGUGAUUUGUUUGCCAAAUUGUGGCAAAUGU ----------....(((..(((((....(((((((((....)))--))).)))...)))))..)))............... ( -20.80, z-score = -1.16, R) >droSec1.super_56 50713 59 - 192059 --------------------GCAGUGGGAUGAGCAGCGUCAGUU--GCUACGUGAUUUGUUUGCCAAAUUGUGGCAAAUGU --------------------........(((((((((....)))--))).))).....((((((((.....)))))))).. ( -18.50, z-score = -1.37, R) >droSim1.chrX 11698710 63 - 17042790 ----------------AUGGGCAGUGGGAUGAGCAGCGUCAGUU--GCUACGUGAUUUGUUUGCCAAAUUGUGGCAAAUGU ----------------............(((((((((....)))--))).))).....((((((((.....)))))))).. ( -18.50, z-score = -0.93, R) >dp4.chrXL_group1a 7508126 65 + 9151740 --------------GAGGCACUGGCGAGGAAUGGACUGGGCAUUU-GCUACGUGAUUUGUUUGCCGAA-UGUGGCAAAUGU --------------......(..((.(....).).)..)((((((-(((((((..............)-)))))))))))) ( -17.64, z-score = -0.94, R) >droPer1.super_53 110941 65 - 525408 --------------GAGGCACUGGCGAGGAAUGGACUGGGCAUUU-GCUACGUGAUUUGUUUGCCGAA-UGUGGCAAAUGU --------------......(..((.(....).).)..)((((((-(((((((..............)-)))))))))))) ( -17.64, z-score = -0.94, R) >droAna3.scaffold_13117 4411075 59 + 5790199 --------------------GAAUCGGAAUGAGCACCGCCAGCU--GCUACGUGAUUUGUUUGCCAAAUUGUGGCAAAUGU --------------------((((((.....((((.(....).)--)))...))))))((((((((.....)))))))).. ( -16.00, z-score = -0.98, R) >droWil1.scaffold_180777 2241000 78 - 4753960 AUGGGAAGUGAGUGUAAAGGGAAUCGGCG-AAGCAACGCCAGUU--CCUACGUGAUUUGUUUGCCAAAUUGUGGCAAAUGU .....((((.(.(((...((((((.((((-......)))).)))--))))))).))))((((((((.....)))))))).. ( -23.90, z-score = -2.06, R) >droMoj3.scaffold_6359 2010207 74 - 4525533 -------UGGCCAAAUGGGGCAGGCGAGUGGAGCAGCGGCAGCUCGGCUACGUGAUUUGUUUGCCAAAUUGUGGCAAAUGU -------..(((((((..(((((((((((.(((((((....)))..)))...).)))))))))))..))).))))...... ( -27.20, z-score = -1.28, R) >consensus ________________GUGGGCAGCGGGAUGAGCAGCGUCAGUU__GCUACGUGAUUUGUUUGCCAAAUUGUGGCAAAUGU ..........................................................((((((((.....)))))))).. ( -9.22 = -9.06 + -0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:45:45 2011