
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,157,644 – 15,157,779 |
| Length | 135 |
| Max. P | 0.690621 |

| Location | 15,157,644 – 15,157,742 |
|---|---|
| Length | 98 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 80.52 |
| Shannon entropy | 0.41684 |
| G+C content | 0.45900 |
| Mean single sequence MFE | -17.95 |
| Consensus MFE | -11.62 |
| Energy contribution | -12.41 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.526220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
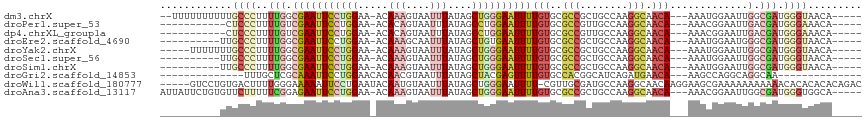
>dm3.chrX 15157644 98 + 22422827 -----UUCUCCGAUUCCUGUUCAUUCUCUCGUGCUGUUACCCAUCGCCAAUUCCAUUUUGUUGCCUUGGCAGCGGCGCACAAAAUUCCCAGCUAUAAAUUACU -----.........................((((((((..(((..((.(((........)))))..))).))))))))......................... ( -16.40, z-score = -0.81, R) >droPer1.super_53 106128 102 + 525408 -UUUUGUACCCGCUCCUUGUUCCUUUUAUCGUGCUGUUUCCCAUCGUCAAUUCCGUUUUGUUGCCUUGGCAACGGCGCACAAAAUUCCAGGCUAUAAAUUACU -.((((((.((((.....))..........((((((((..(((..(.((((........)))))..))).))))))))...........)).))))))..... ( -20.80, z-score = -1.81, R) >dp4.chrXL_group1a 7503514 102 - 9151740 -UUUUGUACCCGCUCCUUGUUCCUUUUAUCGUGCUGUUUCCCAUCGUCAAUUCCGUUUUGUUGCCUUGGCAACGGCGCACAAAAUUCCAGGCUAUAAAUUACU -.((((((.((((.....))..........((((((((..(((..(.((((........)))))..))).))))))))...........)).))))))..... ( -20.80, z-score = -1.81, R) >droEre2.scaffold_4690 6887985 98 - 18748788 -----UUCUCCUAUUCCUGUUCAUUCUCUCGUGCUGUUACCCAUCGCCCAUUCCAUUUUGUUGCCUUGGCAGCGGCGCACAAAAUUCACAGCUAUAAAUUGCU -----......(((..((((..........((((((((..(((..((.((........))..))..))).)))))))).........))))..)))....... ( -17.81, z-score = -1.16, R) >droYak2.chrX 9357547 98 + 21770863 -----UUCUCCGAUUCCUGUUCAUUCUCUCGUGCUGUUACCCAUCGCCAAUUCCAUUUUGUUGCCUUGGCAGCGGCGCACAAAAUUCCCAGCUAUAAAUUACU -----.........................((((((((..(((..((.(((........)))))..))).))))))))......................... ( -16.40, z-score = -0.81, R) >droSec1.super_56 47432 98 + 192059 -----UUCUCCGAUUCCUGUUCAUUCUCUCGUGCUGUUACCCAUCGCCAAUUCCAUUUUGUUGCCUUGGCAGCGGCGCACAAAAUUCCCAGCUAUAAAUUACU -----.........................((((((((..(((..((.(((........)))))..))).))))))))......................... ( -16.40, z-score = -0.81, R) >droSim1.chrX 11695327 98 + 17042790 -----UUCUCCGAUUCCUGUUCAUUCUCUCGUGCUGUUACCCAUCGCCAAUUCCAUUUUGUUGCCUUGGCAGCGGCGCACAAAAUUCCCAGCUAUAAAUUACU -----.........................((((((((..(((..((.(((........)))))..))).))))))))......................... ( -16.40, z-score = -0.81, R) >droGri2.scaffold_14853 6747745 89 - 10151454 ----------CUAUUCCUCUUCUACUUCUUUUGUUGUGC----UUGCCUGCCUGGCUUUGUUCAUCUGAUGCCGUGGCACAAAACUCGUAGCUAUAAAUUACG ----------...........((((...((((((.((..----..))..(((((((.((........)).)))).)))))))))...))))............ ( -14.20, z-score = -0.71, R) >droAna3.scaffold_13117 1376262 103 + 5790199 ACUCUCCUAGCGAUUCCUGUUCAGUCUCCCGUGCUGCCACCCAUCGCCAAUUCCGUUUUGUUGCCUUGGCAGCGGCGCACAAAAUUCCCAGCUAUAAAUUACU .......((((((((.......))))....((((.(((......((.......))....(((((....))))))))))))..........))))......... ( -22.30, z-score = -1.53, R) >consensus _____UUCUCCGAUUCCUGUUCAUUCUCUCGUGCUGUUACCCAUCGCCAAUUCCAUUUUGUUGCCUUGGCAGCGGCGCACAAAAUUCCCAGCUAUAAAUUACU ..............................(((((((...(((..(.((((........)))))..)))..)))))))......................... (-11.62 = -12.41 + 0.79)



| Location | 15,157,673 – 15,157,779 |
|---|---|
| Length | 106 |
| Sequences | 10 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 75.69 |
| Shannon entropy | 0.49058 |
| G+C content | 0.43621 |
| Mean single sequence MFE | -29.92 |
| Consensus MFE | -13.26 |
| Energy contribution | -14.48 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.690621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15157673 106 - 22422827 --UUUUUUUUUUGCCCUUUUGGCGAAUUCCUGGAA-ACAAAGUAAUUUAUAGCUGGGAAUUUUGUGCGCCGCUGCCAAGGCAACA---AAAUGGAAUUGGCGAUGGGUAACA----- --........((((((...(.((.((((((((...-.))...................(((((((..(((........))).)))---)))))))))).)).).))))))..----- ( -32.60, z-score = -1.64, R) >droPer1.super_53 106161 97 - 525408 -----------CUCCCUUUUGUCGAAUUCCUGGAA-ACACAGUAAUUUAUAGCCUGGAAUUUUGUGCGCCGUUGCCAAGGCAACA---AAACGGAAUUGACGAUGGGAAACA----- -----------.((((..(((((.((((((.((..-.(((((.(((((........))))))))))..))(((((....))))).---....))))))))))).))))....----- ( -33.90, z-score = -3.91, R) >dp4.chrXL_group1a 7503547 97 + 9151740 -----------CUCCCUUUUGUCGAAUUCCUGGAA-ACACAGUAAUUUAUAGCCUGGAAUUUUGUGCGCCGUUGCCAAGGCAACA---AAACGGAAUUGACGAUGGGAAACA----- -----------.((((..(((((.((((((.((..-.(((((.(((((........))))))))))..))(((((....))))).---....))))))))))).))))....----- ( -33.90, z-score = -3.91, R) >droEre2.scaffold_4690 6888014 98 + 18748788 ----------UUGCCCUUUUGGCGAAUUCCUGGAA-ACAAAGCAAUUUAUAGCUGUGAAUUUUGUGCGCCGCUGCCAAGGCAACA---AAAUGGAAUGGGCGAUGGGUAACA----- ----------((((((...(.((..(((((((...-.)).(((........)))....(((((((..(((........))).)))---)))))))))..)).).))))))..----- ( -31.30, z-score = -1.39, R) >droYak2.chrX 9357576 103 - 21770863 -----UUUUUUUGCCCUUUUGGCGAAUUCCUGGAA-ACAAAGUAAUUUAUAGCUGGGAAUUUUGUGCGCCGCUGCCAAGGCAACA---AAAUGGAAUUGGCGAUGGGUAACA----- -----.....((((((...(.((.((((((((...-.))...................(((((((..(((........))).)))---)))))))))).)).).))))))..----- ( -32.60, z-score = -1.81, R) >droSec1.super_56 47461 98 - 192059 ----------UUGCCCUUUUGGCGAAUUCCUGGAA-ACAAAGUAAUUUAUAGCUGGGAAUUUUGUGCGCCGCUGCCAAGGCAACA---AAAUGGAAUUGGCGAUGGGUAACA----- ----------((((((...(.((.((((((((...-.))...................(((((((..(((........))).)))---)))))))))).)).).))))))..----- ( -32.40, z-score = -1.97, R) >droSim1.chrX 11695356 98 - 17042790 ----------UUGCCCUUUUGGCGAAUUCCUGGAA-ACAAAGUAAUUUAUAGCUGGGAAUUUUGUGCGCCGCUGCCAAGGCAACA---AAAUGGAAUUGGCGAUGGGUAACA----- ----------((((((...(.((.((((((((...-.))...................(((((((..(((........))).)))---)))))))))).)).).))))))..----- ( -32.40, z-score = -1.97, R) >droGri2.scaffold_14853 6747774 86 + 10151454 --------------UUUGCUCGCAAAUUCCUGGAACACAACGUAAUUUAUAGCUACGAGUUUUGUGCCACGGCAUCAGAUGAACA---AAGCCAGGCAGGCAA-------------- --------------.......((.....(((((.......((((.........)))).((((.((((....)))).)))).....---...)))))...))..-------------- ( -18.60, z-score = 0.04, R) >droWil1.scaffold_180777 2505580 111 + 4753960 -----GUCCUGUGACUUUUGGGAAAAAUUCCUGAAUACAAUGUAAUUUAUAGCUGGGAAUUUU-CGUUGCGAUGCCAAGGCAACAAGGAAGCGAAAAAAAAAAACACACACACAGAC -----...(((((...((..(((.....)))..))........................((((-((((.(..(((....)))....)..)))))))).............))))).. ( -21.80, z-score = -0.62, R) >droAna3.scaffold_13117 1376296 108 - 5790199 AUUAUUCUGUGUUCUUUUUCGGAGAAUUCCUGGAA-ACAAAGUAAUUUAUAGCUGGGAAUUUUGUGCGCCGCUGCCAAGGCAACA---AAACGGAAUUGGCGAUGGGUGGCA----- ...................((.((((((((..(..-..(((....)))....)..)))))))).)).(((((((((((..(....---.....)..)))))....)))))).----- ( -29.70, z-score = -0.69, R) >consensus __________UUGCCCUUUUGGCGAAUUCCUGGAA_ACAAAGUAAUUUAUAGCUGGGAAUUUUGUGCGCCGCUGCCAAGGCAACA___AAACGGAAUUGGCGAUGGGUAACA_____ ............((((..(((.(((((((((((.....(((....)))....)))))))))).....(((........))).................).))).))))......... (-13.26 = -14.48 + 1.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:45:43 2011