| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,156,127 – 15,156,230 |
| Length | 103 |
| Max. P | 0.990362 |

| Location | 15,156,127 – 15,156,230 |
|---|---|
| Length | 103 |
| Sequences | 11 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 70.15 |
| Shannon entropy | 0.64571 |
| G+C content | 0.40227 |
| Mean single sequence MFE | -21.40 |
| Consensus MFE | -12.78 |
| Energy contribution | -13.26 |
| Covariance contribution | 0.49 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.990362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15156127 103 - 22422827 GAUAUUCGCCCACUGCUCACCCUCCCAAAUGGCCGAUAAAUUGAUUUAUGUGCUUGUUUGUCAGUUGAACUGCAACU--UGUGCAUAAAUUUCAAAUUGCCAUUG------- .......((.....))...........((((((.(((.....((((((((..(..(((.(.(((.....))))))).--.)..))))))))....))))))))).------- ( -23.30, z-score = -2.38, R) >droAna3.scaffold_13117 1374609 91 - 5790199 ------UACGGCCCGCUCCUCUCCCCA---GGCCGAUAAAUUGAUUUAUGUGCUUGUUUGUCAGUUGAACUGCAACU--UGUGCAUAAAUUUCAAAUUGCCA---------- ------..(((((..............---))))).......((((((((..(..(((.(.(((.....))))))).--.)..))))))))...........---------- ( -23.44, z-score = -2.56, R) >droEre2.scaffold_4690 6886444 103 + 18748788 GAUAUUCGCCCACUGCUCAUUCUCCCAAAUGGCCGAUAAAUUGAUUUAUGUGCUUGUUUGUCAGUUGAACUGCAACU--UGUGCAUAAAUUUCAAAUUGCCAUUG------- .......((.....))...........((((((.(((.....((((((((..(..(((.(.(((.....))))))).--.)..))))))))....))))))))).------- ( -23.30, z-score = -2.16, R) >droYak2.chrX 9355935 91 - 21770863 ------------CUGCUCACACUCCCAAAUGGCCGAUAAAUUGAUUUAUGUGCUUGUUUGUCAGUUGAACUGCAACU--UGUGCAUAAAUUUCAAAUUGCCAUUG------- ------------...............((((((.(((.....((((((((..(..(((.(.(((.....))))))).--.)..))))))))....))))))))).------- ( -23.20, z-score = -2.72, R) >droSec1.super_56 45939 103 - 192059 GAUAUUCGCCCACUGCUCACCCUCCCAAAUGGCCGAUAAAUUGAUUUAUGUGCUUGUUUGUCAGUUGAACUGCAACU--UGUGCAUAAAUUUCAAAUUGCCAUUG------- .......((.....))...........((((((.(((.....((((((((..(..(((.(.(((.....))))))).--.)..))))))))....))))))))).------- ( -23.30, z-score = -2.38, R) >droSim1.chrX 11693848 103 - 17042790 GAUAUUCGCCCACUGCUCACCCUCCCAAAUGGCCGAUAAAUUGAUUUAUGUGCUUGUUUGUCAGUUGAACUGCAACU--UGUGCAUAAAUUUCAAAUUGCCAUUG------- .......((.....))...........((((((.(((.....((((((((..(..(((.(.(((.....))))))).--.)..))))))))....))))))))).------- ( -23.30, z-score = -2.38, R) >droWil1.scaffold_180777 2502456 103 + 4753960 CCUCCCCGCCCACACCUCCUUCAACAACCAUGUCGAUGAAUUGAUUUAUGGGCUUGUUUGUCAGUUGAACUGCAACU--UGUGCAUAAAUUUCAAAAUGGCAAGA------- .......(((..........((.(((....))).))(((...((((((((.((..(((.(.(((.....))))))).--.)).)))))))))))....)))....------- ( -20.50, z-score = -0.24, R) >droMoj3.scaffold_6359 2001185 93 - 4525533 ---------UGUGUGCUCAUGUCAUU--UUAGUCGAUGAAUUGAUUUAUGUGCUUGUUUGUCAGCUGACCUGCAACU--UGUGCAUAAAUUUCAAAU------UUCACAUUG ---------.(((((.(((..(((((--......)))))..)))((((((..(((((..(((....)))..))))..--.)..))))))........------..))))).. ( -20.60, z-score = -0.82, R) >droGri2.scaffold_14853 6745372 96 + 10151454 ---------------CAUGUUUGACUG-UUAGUCGAUGAAUUGAUUUAUGUGCUUGUUUCUCAGUUGACCUGCAACUUGUGUGCAUAAAUUUCAAAUAGCUGCCAUGCAUUG ---------------((((.(((((..-...)))))(((...((((((((..(..(((...(((.....))).)))....)..))))))))))).........))))..... ( -17.90, z-score = 0.59, R) >dp4.chrXL_group1a 7497106 102 + 9151740 -------CAACUUUGCCACUCCCCUGCUCUUGCCGAUGAAUUGAUUUAUGUGCUUGUUUGUCAGGUGAACUGCAACU--UGUGCAUAAAUUUAAAAUUGUAAUUACCAGUG- -------.........((((.....((....))((((.....((((((((..(..(((.(.(((.....))))))).--.)..))))))))....))))........))))- ( -20.80, z-score = -1.50, R) >droPer1.super_53 97538 86 - 525408 ---------CACUUGCCACU--CCUGCUCU--GCGAUGAAU--GAUUAUGUGC-UGUUUGUCAGGUG-ACUGCA--C--UGUGCAUAAAUUAAAUGU----AUUACCAGUG- ---------((((((.((..--...((...--(((((((..--..)))).)))-.)).)).))))))-((((..--.--.((((((.......))))----))...)))).- ( -15.80, z-score = 0.81, R) >consensus _______GCCCACUGCUCAUCCUCCCAAAUGGCCGAUAAAUUGAUUUAUGUGCUUGUUUGUCAGUUGAACUGCAACU__UGUGCAUAAAUUUCAAAUUGCCAUUG_______ ..............................(((.........((((((((..(....(((.(((.....)))))).....)..)))))))).......)))........... (-12.78 = -13.26 + 0.49)


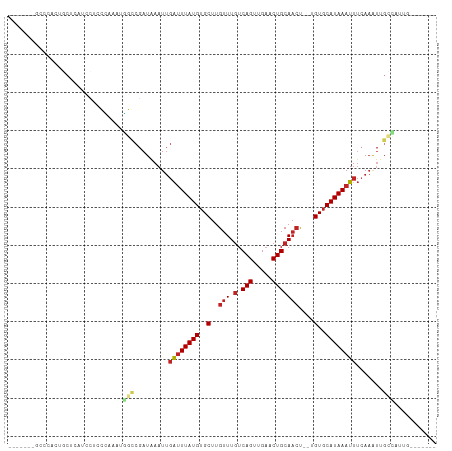
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:45:42 2011