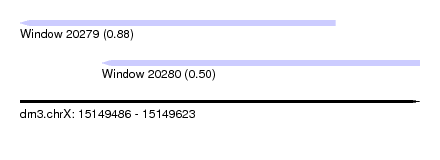
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,149,486 – 15,149,623 |
| Length | 137 |
| Max. P | 0.878327 |

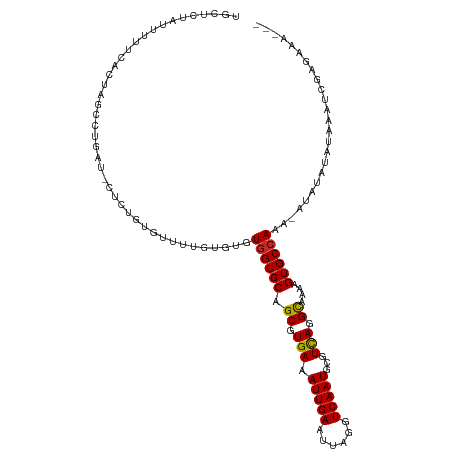
| Location | 15,149,486 – 15,149,594 |
|---|---|
| Length | 108 |
| Sequences | 10 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 71.09 |
| Shannon entropy | 0.62310 |
| G+C content | 0.41173 |
| Mean single sequence MFE | -25.18 |
| Consensus MFE | -16.27 |
| Energy contribution | -16.04 |
| Covariance contribution | -0.23 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.63 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.878327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15149486 108 - 22422827 UGCUCUAUUUUUCACUAGCCUGAU-CUCUGUGUUUUGUGUGUGGCGCAGCGUGAAAUUGAAUUAGGUCAAUGCGUCAGGCAGAAGUGCCAAA-A--UAUAUAAAUCGAGAAA--- .......................(-(((.(...(((((((((((((((...(((..(......)..))).)))))).((((....))))...-)--)))))))).)))))..--- ( -25.60, z-score = -0.23, R) >droSim1.chrX 11687326 110 - 17042790 UGCUCUAUUUCUCACUAGCCUGAU-CUCUGUGUUUUGUGUGUGGCGCAGCGUGAAAUUGAAUUAGGUCAAUGCGUCAGGCAAAAGUGCCAAA-AUAUAUAUAAAUCGAGAAA--- .......((((((.(......)..-......(.(((((((((((((((...(((..(......)..))).)))))).((((....))))...-..))))))))).)))))))--- ( -26.30, z-score = -0.79, R) >droSec1.super_56 39388 110 - 192059 UGCUCUAUUUCUCACUAGCCUGAU-CUCUGUGUUUUGUGUGUGGCGCAGCGUGAAAUUGAAUUAGGUCAAUGCGUCAGGCAAAAGUGCCAAA-AUGUAUAUAAAUCGAGAAA--- .......((((((....(((((((-.((...((((..(((........)))..)))).)))))))))..((((((..((((....))))...-)))))).......))))))--- ( -27.80, z-score = -1.13, R) >droYak2.chrX 9348974 110 - 21770863 UGCUCUAUUUUUCACUACCCUGAU-CUCUGUGUUUUGUGUGUGGCGCAGCGUGAAAUUGAAUUAGGUCAAUGCGUCAGGCAAAAGUGCCAAA-AUAUAUAUAAAUCGAGAAA--- .......................(-(((.(...(((((((((((((((...(((..(......)..))).)))))).((((....))))...-..))))))))).)))))..--- ( -25.60, z-score = -0.88, R) >droEre2.scaffold_4690 6880145 108 + 18748788 UGCUCUAUUUUUCACUACCCUGAU-CUCUGUGUUUUGUGUGUUGCGCAGCGUGAAAUUGAAUUAGGUCAAUGCGUCAGGCAAAAGUGCCAAA-A--UAUAUAAAUCGAGAAA--- .......................(-(((.(...(((((((((((((((...(((..(......)..))).)))))..((((....))))..)-)--)))))))).)))))..--- ( -23.20, z-score = -0.45, R) >droAna3.scaffold_13117 1368267 100 - 5790199 ------------UGCUCCUUCGGCGCUCCGUGUCCUGUGUGUGGCGCAGCGUGAAAUUGAAUUAGGUCAAUGCGUCAGGCAAGAGUGCCAAAUGUGUAUAGAAAUCGGGAAA--- ------------...((((..(((((((.((((((.....).))))).((.(((.(((((......)))))...))).))..)))))))...........(....)))))..--- ( -31.50, z-score = -1.15, R) >droWil1.scaffold_180777 2257284 91 - 4753960 CAUAUUGUGUUGCGUUGUGCUGUG-CACUGUUGUUUGUGUUUGGCGCAGCGUGAAAUUGAAUUAGGUCAAUGCGUUAAGUAAAAGUGCCAAA----------------------- .........(..((((((((((.(-(((........)))).))))))))))..)..........((.((.(((.....)))....))))...----------------------- ( -26.00, z-score = -1.17, R) >droVir3.scaffold_13042 2087125 110 - 5191987 -----UUCUUUUUUCUUAGAUUCCUUUUCCCUGCCUGUGUCUGGCGCAGCGUGAAAUUGAAUUAGGUCAAUGCGUCAAGUAAAAGUGCCAAAUAUAAAAUGACAACAACAAGAAG -----.......(((((.(((((..(((((((((((......)).)))).).))))..))))).((.((.(((.....)))....))))....................))))). ( -19.00, z-score = 0.20, R) >droMoj3.scaffold_6359 1988613 97 - 4525533 ---------CUCCUUUUGCCACUUGCUGCGUUGUUUGCGUCUGGCGCAGCGUGAAAUUGAAUUAGGUCAAUGCGUCAAGUAAAAGUGCCAAAUACAAAACAAGUAA--------- ---------.....((((.((((((((((((((........))))))))).(((.(((((......)))))...))).....))))).))))..............--------- ( -24.00, z-score = -0.31, R) >droGri2.scaffold_14853 3396452 103 - 10151454 ----------GUUUCUU--UUCUUCUUGCGUUGUUUGUGUGUGGCGCAGCGUGAAAUUGAAUUAGGUCAAUGCGUCAAGUAAAAGUGCCAAAUAUAAAAUGACAACUACAGUGUU ----------.......--..........(((((((((((.((((((.((.(((.(((((......)))))...))).))....)))))).)))))....))))))......... ( -22.80, z-score = -0.38, R) >consensus UGCUCUAUUUUUCACUAGCCUGAU_CUCUGUGUUUUGUGUGUGGCGCAGCGUGAAAUUGAAUUAGGUCAAUGCGUCAGGCAAAAGUGCCAAA_AUAUAUAUAAAUCGAGAAA___ .........................................((((((.((.(((.(((((......)))))...))).))....))))))......................... (-16.27 = -16.04 + -0.23)

| Location | 15,149,514 – 15,149,623 |
|---|---|
| Length | 109 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.59 |
| Shannon entropy | 0.59513 |
| G+C content | 0.44460 |
| Mean single sequence MFE | -22.86 |
| Consensus MFE | -13.96 |
| Energy contribution | -13.80 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.15 |
| Mean z-score | -0.36 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chrX 15149514 109 - 22422827 --------CGUUUUUUUUUUUUGCCUCCCUAUUAUUUUGCU-CUAUUUUUCACUAGCCUGAU--CUCUGUGUUUUGUGUGUGGCGCAGCGUGAAAUUGAAUUAGGUCAAUGCGUCAGGCA --------.................................-.............(((((((--(.(((((((........))))))).)....(((((......)))))..))))))). ( -20.80, z-score = 0.17, R) >droSim1.chrX 11687356 106 - 17042790 -----------GCCUUUUAUUCGCCUCCCUAUUAUUUUGCU-CUAUUUCUCACUAGCCUGAU--CUCUGUGUUUUGUGUGUGGCGCAGCGUGAAAUUGAAUUAGGUCAAUGCGUCAGGCA -----------((((......(((.......((((.((((.-((((....(((.((((....--....).)))..))).)))).)))).)))).(((((......))))))))..)))). ( -22.00, z-score = 0.16, R) >droSec1.super_56 39418 106 - 192059 -----------GCCUUUUAUUCGCCUCCCUAUUAUUUUGCU-CUAUUUCUCACUAGCCUGAU--CUCUGUGUUUUGUGUGUGGCGCAGCGUGAAAUUGAAUUAGGUCAAUGCGUCAGGCA -----------((((......(((.......((((.((((.-((((....(((.((((....--....).)))..))).)))).)))).)))).(((((......))))))))..)))). ( -22.00, z-score = 0.16, R) >droYak2.chrX 9349004 103 - 21770863 -----------GCCUUUUAUUCG---CCUAACUAUUUUGCU-CUAUUUUUCACUACCCUGAU--CUCUGUGUUUUGUGUGUGGCGCAGCGUGAAAUUGAAUUAGGUCAAUGCGUCAGGCA -----------((((..((((.(---(((((..((((..(.-.......(((......))).--..(((((((........))))))).)..))))....)))))).))))....)))). ( -23.90, z-score = -1.01, R) >droEre2.scaffold_4690 6880173 106 + 18748788 -----------GCCUUCUAUUCGACUCCCCAUUAUUUUGCU-CUAUUUUUCACUACCCUGAU--CUCUGUGUUUUGUGUGUUGCGCAGCGUGAAAUUGAAUUAGGUCAAUGCGUCAGGCA -----------((((.(((((.((((.......((((..(.-.......(((......))).--..((((((..........)))))).)..)))).......)))))))).)..)))). ( -19.44, z-score = 0.08, R) >droAna3.scaffold_13117 1368298 82 - 5790199 ------------------------------------GUUUC-UUGUUCCUUGCUCCUUCGGC-GCUCCGUGUCCUGUGUGUGGCGCAGCGUGAAAUUGAAUUAGGUCAAUGCGUCAGGCA ------------------------------------.....-..................((-((..((..(...)..))..)))).((.(((.(((((......)))))...))).)). ( -20.10, z-score = 0.59, R) >droWil1.scaffold_180777 2257295 117 - 4753960 GUCCAUUCUUCUCGCUUUUCACAUUUUCCCAUUUUUCCAUA-UUGUGUUGCGUUGUGCUGU--GCACUGUUGUUUGUGUUUGGCGCAGCGUGAAAUUGAAUUAGGUCAAUGCGUUAAGUA .......(((..(((....((((..................-.))))(..((((((((((.--((((........)))).))))))))))..).(((((......))))))))..))).. ( -30.41, z-score = -2.28, R) >droMoj3.scaffold_6359 1988638 96 - 4525533 ------------------------GUUUUCGUUGUCGCUUUACCAUUUCUCCUUUUGCCACUUGCUGCGUUGUUUGCGUCUGGCGCAGCGUGAAAUUGAAUUAGGUCAAUGCGUCAAGUA ------------------------.......(((.(((((.(((..(((...(((..(.....(((((((((........))))))))))..)))..)))...))).)).))).)))... ( -24.20, z-score = -0.74, R) >consensus ___________GCCUUUUAUUCG_CUCCCCAUUAUUUUGCU_CUAUUUCUCACUACCCUGAU__CUCUGUGUUUUGUGUGUGGCGCAGCGUGAAAUUGAAUUAGGUCAAUGCGUCAGGCA .............................................................................((.(((((((...(((..(......)..))).))))))).)). (-13.96 = -13.80 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:45:41 2011