
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,147,719 – 15,147,819 |
| Length | 100 |
| Max. P | 0.853062 |

| Location | 15,147,719 – 15,147,819 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 56.66 |
| Shannon entropy | 0.78019 |
| G+C content | 0.41259 |
| Mean single sequence MFE | -24.35 |
| Consensus MFE | -5.92 |
| Energy contribution | -5.92 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.53 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.24 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.853062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
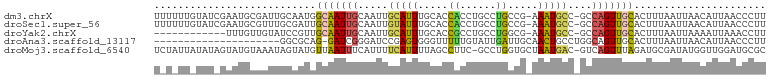
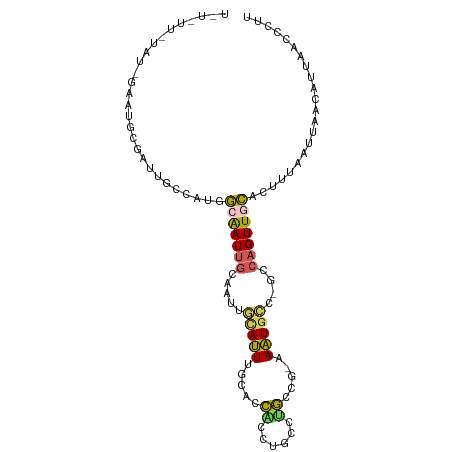
>dm3.chrX 15147719 100 - 22422827 UUUUUUGUAUCGAAUGCGAUUGCAAUGGCAAUUGCAAUUGCAUUUGCACCACCUGCCUGCCG-AAAUGCC-GCCAGUUGCACUUUAAUUAACAUUAACCCUU .....(((..((((((((((((((((....)))))))))))))))).......((((((.((-......)-).)))..))).........)))......... ( -26.40, z-score = -2.59, R) >droSec1.super_56 37645 100 - 192059 UUUUUUGUAUCGAAUGCGUUUGCGAUUGCAAUUGCAAUUGUAUUUGCACCACCUGCCUGCCG-AAAUGCC-GCCAGUUGCACUUUAAUUAACAUUAACCCUU .....(((......((((..(((((((((....)))))))))..)))).....((((((.((-......)-).)))..))).........)))......... ( -22.10, z-score = -1.87, R) >droYak2.chrX 9347155 88 - 21770863 ------------UUUGUUGUAUCCGUUGCAAUUGCAAUUGCAUUUGCACCGCCUGCCUGGCG-AAAUGCC-GCCAGUUGCACUUUAAUUAAAAUUAAACCUU ------------......(((..((.(((((.(((....))).))))).))..)))((((((-......)-))))).......................... ( -23.30, z-score = -2.11, R) >droAna3.scaffold_13117 1366756 80 - 5790199 ---------------------GGCGCAG-GAUCGGGAUCCGAGUGGGUUUUUGUAUUGAUUGCAACUGCCUGGCAGUUGCACUUUAAUUAACAUUAACCCUU ---------------------.((...(-(((....))))..))(((((..((((((((.(((((((((...)))))))))..)))))..)))..))))).. ( -30.10, z-score = -3.59, R) >droMoj3.scaffold_6540 16547943 100 - 34148556 UCUAUUAUAUAGUAUGUAAAUAGUAUGUUAAUUUCAUUUUCAUUUUAGCCUUC-GCCUGGUGCUAAUGAC-GUCAGUUUAGAUGCGAUAUGGUUGGAUGCGC ((((((((((.((((.(((((.(.((((((..............(((((((..-....)).)))))))))-))).))))).)))).)))))).))))..... ( -19.83, z-score = 0.03, R) >consensus U_U_UU_UAU_GAAUGCGAUUGCCAUGGCAAUUGCAAUUGCAUUUGCACCACCUGCCUGCCG_AAAUGCC_GCCAGUUGCACUUUAAUUAACAUUAACCCUU ...........................(((((((.....((((((....((......))....))))))....)))))))...................... ( -5.92 = -5.92 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:45:39 2011