| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,133,404 – 15,133,522 |
| Length | 118 |
| Max. P | 0.983437 |

| Location | 15,133,404 – 15,133,522 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 78.94 |
| Shannon entropy | 0.40299 |
| G+C content | 0.47769 |
| Mean single sequence MFE | -31.03 |
| Consensus MFE | -22.64 |
| Energy contribution | -23.40 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.983437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15133404 118 - 22422827 CCCACUUAUCCAUAUCCA-UGCCACAUCCUGCUGCACAGAUUGCCACCUGGCUGGCUGCCCAUCAAUUCAAAUGCC--AUCCACCGUUUGAAUUGAUGAUGUCGAUGAUGGUAUCGCCCCC .................(-(((((((((........(((...(((....)))...)))..((((((((((((((..--......)))))))))))))).....)))).))))))....... ( -34.40, z-score = -2.65, R) >droMoj3.scaffold_6359 2594215 111 + 4525533 --------UUCUUAUUGAGAGGAAUAUUUUACAA-AUAGAUUAAUAAAUGCUAAAGGGUUAAUCAAUGCAACUAUCGAAUCAAGUGCUGGCACUCAAAACUGCAAAGAU-GUAGGACAAUU --------......(((((...............-...(((((((...(.....)..)))))))..(((.(((.........)))....))))))))..(((((....)-))))....... ( -15.90, z-score = 0.47, R) >droEre2.scaffold_4690 6864454 118 + 18748788 CCCACUUAUACAUAUCCA-UUCCACAUCCUGCUGCACAGAUUGCCACCUGGCUGGCUGCCCAUCAAUUCAAAUGCC--AUCCACCGUUUGAAUUGAUGAUGUCGAUGAUGGUAUCGCCCCC .......((((.((((..-.((.(((((........(((...(((....)))...)))...(((((((((((((..--......)))))))))))))))))).)).))))))))....... ( -30.80, z-score = -2.03, R) >droYak2.chrX 9333118 118 - 21770863 CCCACUUAUCCAUAUCCA-UUCCACGUCCUGCAGCACAGAUUGCCACCUGGCUGGCUGCCCAUCAAUUCGAAUGCC--AUCCACCGUUUGAAUUGAUGAUGUCGAUGAUGGUAUCGCUCCU .(((.(((((.(((((..-...........(((((.((....(((....))))))))))..(((((((((((((..--......)))))))))))))))))).)))))))).......... ( -35.50, z-score = -2.88, R) >droSec1.super_56 22473 118 - 192059 CCCACUUAUCCAUAUCAA-UGCCACAUCCUGCUGCACAGAUUGCCACCUGGCUGGCUGCCCAUCAAUUCAAAUGCC--AUCCACCGUUUGAAUUGAUGAUGUCGAUGAUGGUAUCGCCCCC .(((.(((((.(((((..-.((((....(((.....)))...(((....))))))).....(((((((((((((..--......)))))))))))))))))).)))))))).......... ( -35.20, z-score = -2.93, R) >droSim1.chrX 11670909 118 - 17042790 CCCACUUAUCCAUAUCCA-UGCCACAUCCUGCUGCACAGAUUGCCACCUGGCUGGCUGCCCAUCAAUUCAAAUGCC--AUCCACCGUUUGAAUUGAUGAUGUCGAUGAUGGUAUCGCCCCC .................(-(((((((((........(((...(((....)))...)))..((((((((((((((..--......)))))))))))))).....)))).))))))....... ( -34.40, z-score = -2.65, R) >consensus CCCACUUAUCCAUAUCCA_UGCCACAUCCUGCUGCACAGAUUGCCACCUGGCUGGCUGCCCAUCAAUUCAAAUGCC__AUCCACCGUUUGAAUUGAUGAUGUCGAUGAUGGUAUCGCCCCC ...................(((((((((........(((...(((....)))...)))..((((((((((((((..........)))))))))))))).....)))).)))))........ (-22.64 = -23.40 + 0.76)

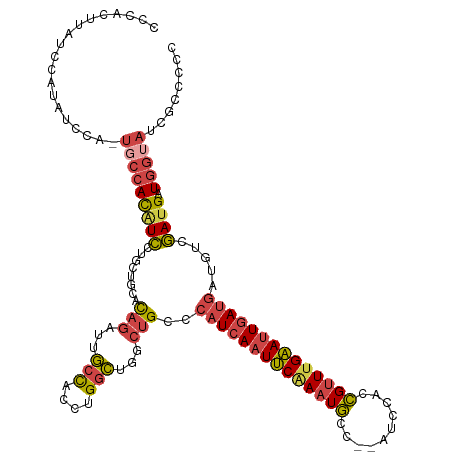
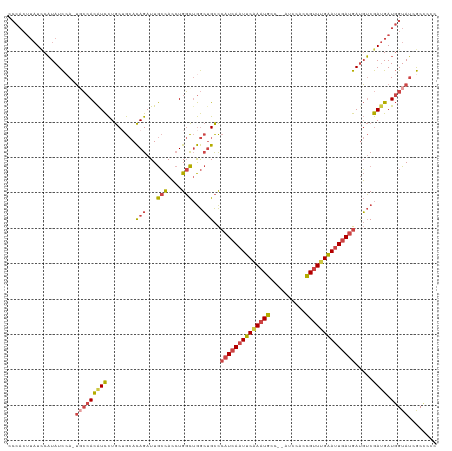
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:45:38 2011