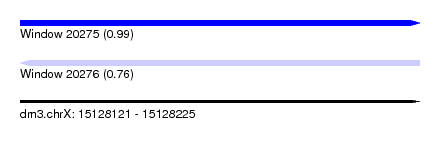
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,128,121 – 15,128,225 |
| Length | 104 |
| Max. P | 0.988933 |

| Location | 15,128,121 – 15,128,225 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 76.66 |
| Shannon entropy | 0.46038 |
| G+C content | 0.56451 |
| Mean single sequence MFE | -35.93 |
| Consensus MFE | -24.11 |
| Energy contribution | -26.09 |
| Covariance contribution | 1.98 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.988933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15128121 104 + 22422827 ----GGCAUAAUCUUUGUGCCAACGCCAGCCGCCGAC-------GUCGGCGUCAACGUAUCCCCAGGCUGCCAGUUUACAAUUUUAGCAGCCGACACCGAGUGAUGGCAACUUGG ----((((((.....))))))...((((..(((((..-------(((.(((....))).......((((((.((.........)).)))))))))..)).))).))))....... ( -35.90, z-score = -2.11, R) >droSim1.chrX 11665773 104 + 17042790 ----GGCAUAAUCUUUGUGCCAACGCCAGCCGCCGAC-------GUCGGCGUCAACGUAUCCCCAGGCUGCCAGUUUACAAUUUUAGCAGCCGACACCGAGUGAUGGCAACUUGG ----((((((.....))))))...((((..(((((..-------(((.(((....))).......((((((.((.........)).)))))))))..)).))).))))....... ( -35.90, z-score = -2.11, R) >droSec1.super_56 17111 104 + 192059 ----GGCAUAAUCUUUGUGCCAACGCCAGCCGCCGAC-------GUCGGCGUCAACGUAUCCCCAGGCUGCCAGUUUACAAUUUUAGCAGCCGACACCGAGUGAUGGCAACUUGG ----((((((.....))))))...((((..(((((..-------(((.(((....))).......((((((.((.........)).)))))))))..)).))).))))....... ( -35.90, z-score = -2.11, R) >droYak2.chrX 9328075 104 + 21770863 ----GGCAUAAUCUUUGUGCCAACGCCAGCCGCCGAC-------GUCGGCGUCAUCGUAUCCCCAGGCUGCCAGUCUAAAAUUUUAGCAGCCCACAUCGAGUGAAGGCAGCCUGG ----((((((.....)))))).(((...(.(((((..-------..))))).)..)))....((((((((((.(.((((....)))))....(((.....)))..)))))))))) ( -42.80, z-score = -4.49, R) >droEre2.scaffold_4690 6859472 111 - 18748788 ----GGCAUAAUCUUUGUGCCAACGCCAGCCGCCAGCCGCCGACGUCGGCGUCAACGUAUCCCCAGGCUGCCAGUUUACAAUUUUGGCAGCCGACACCGAGUGAUGGCAAGUUGG ----((((((.....))))))....(((((.((((..(((((..(((.(((....))).......(((((((((.........))))))))))))..)).))).))))..))))) ( -46.80, z-score = -3.56, R) >droAna3.scaffold_13117 1348835 107 + 5790199 GAGGGGCGUAAUCUUUGUGCCAACGCCAGUCGCCAGC-------GUUGGCGUCAACUUAUCCCCAGGCUGCCAGUUUACAAUUUUG-CAGCCGGCCGACAGGGCUGGCAAGUUGG ..((((.((((...((((((((((((.........))-------))))))).))).))))))))((((.....)))).((((((..-..(((((((.....))))))))))))). ( -41.10, z-score = -0.97, R) >droGri2.scaffold_14853 3371622 94 + 10151454 --------CUCUUUCUCAGUCAUCGCCCACCAUCGUCG--CAUAAUCCCAGGCAACGUGACCC--AGCUGACAGUUUACAAUUUGCACAGUAUUCGACAAAUGCUG--------- --------..........((((..((.......(((.(--(..........)).)))......--.))))))...............(((((((.....)))))))--------- ( -13.14, z-score = 0.58, R) >consensus ____GGCAUAAUCUUUGUGCCAACGCCAGCCGCCGAC_______GUCGGCGUCAACGUAUCCCCAGGCUGCCAGUUUACAAUUUUAGCAGCCGACACCGAGUGAUGGCAACUUGG ....((((((.....))))))...(((.(.(((((((.......))))))).)............((((((...............)))))).............)))....... (-24.11 = -26.09 + 1.98)

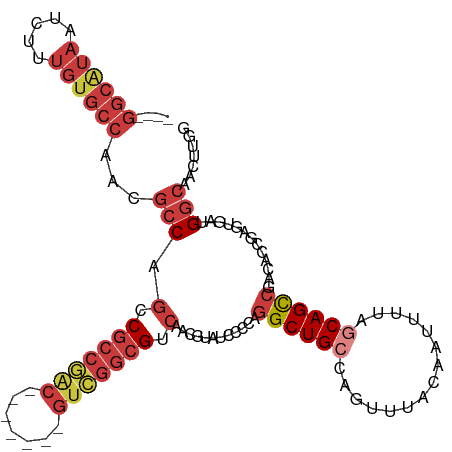
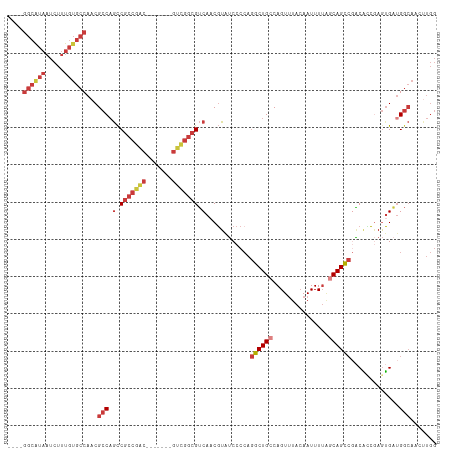
| Location | 15,128,121 – 15,128,225 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 76.66 |
| Shannon entropy | 0.46038 |
| G+C content | 0.56451 |
| Mean single sequence MFE | -38.98 |
| Consensus MFE | -21.82 |
| Energy contribution | -23.61 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.758472 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15128121 104 - 22422827 CCAAGUUGCCAUCACUCGGUGUCGGCUGCUAAAAUUGUAAACUGGCAGCCUGGGGAUACGUUGACGCCGAC-------GUCGGCGGCUGGCGUUGGCACAAAGAUUAUGCC---- ...(((((((...((((((((((((((((((...........)))))))).(......)...)))))))).-------)).)))))))(((((...(.....)...)))))---- ( -40.50, z-score = -1.82, R) >droSim1.chrX 11665773 104 - 17042790 CCAAGUUGCCAUCACUCGGUGUCGGCUGCUAAAAUUGUAAACUGGCAGCCUGGGGAUACGUUGACGCCGAC-------GUCGGCGGCUGGCGUUGGCACAAAGAUUAUGCC---- ...(((((((...((((((((((((((((((...........)))))))).(......)...)))))))).-------)).)))))))(((((...(.....)...)))))---- ( -40.50, z-score = -1.82, R) >droSec1.super_56 17111 104 - 192059 CCAAGUUGCCAUCACUCGGUGUCGGCUGCUAAAAUUGUAAACUGGCAGCCUGGGGAUACGUUGACGCCGAC-------GUCGGCGGCUGGCGUUGGCACAAAGAUUAUGCC---- ...(((((((...((((((((((((((((((...........)))))))).(......)...)))))))).-------)).)))))))(((((...(.....)...)))))---- ( -40.50, z-score = -1.82, R) >droYak2.chrX 9328075 104 - 21770863 CCAGGCUGCCUUCACUCGAUGUGGGCUGCUAAAAUUUUAGACUGGCAGCCUGGGGAUACGAUGACGCCGAC-------GUCGGCGGCUGGCGUUGGCACAAAGAUUAUGCC---- (((((((((((((((.....)))))...((((....))))...))))))))))............((((((-------(((((...)))))))))))..............---- ( -43.90, z-score = -3.04, R) >droEre2.scaffold_4690 6859472 111 + 18748788 CCAACUUGCCAUCACUCGGUGUCGGCUGCCAAAAUUGUAAACUGGCAGCCUGGGGAUACGUUGACGCCGACGUCGGCGGCUGGCGGCUGGCGUUGGCACAAAGAUUAUGCC---- ......(((((....((((((((((((((((...........)))))))).(......)...))))))))(((((((.(....).))))))).))))).............---- ( -47.00, z-score = -2.09, R) >droAna3.scaffold_13117 1348835 107 - 5790199 CCAACUUGCCAGCCCUGUCGGCCGGCUG-CAAAAUUGUAAACUGGCAGCCUGGGGAUAAGUUGACGCCAAC-------GCUGGCGACUGGCGUUGGCACAAAGAUUACGCCCCUC .(((((((((.(((.....))).(((((-(..............))))))...)).)))))))..((((((-------((..(...)..)))))))).................. ( -41.24, z-score = -1.48, R) >droGri2.scaffold_14853 3371622 94 - 10151454 ---------CAGCAUUUGUCGAAUACUGUGCAAAUUGUAAACUGUCAGCU--GGGUCACGUUGCCUGGGAUUAUG--CGACGAUGGUGGGCGAUGACUGAGAAAGAG-------- ---------...((.(..(((..((((((.(...(((((....(((..(.--.(((......)))..))))..))--))).)))))))..)))..).))........-------- ( -19.20, z-score = 1.53, R) >consensus CCAAGUUGCCAUCACUCGGUGUCGGCUGCUAAAAUUGUAAACUGGCAGCCUGGGGAUACGUUGACGCCGAC_______GUCGGCGGCUGGCGUUGGCACAAAGAUUAUGCC____ ......(((((............((((((((...........)))))))).......(((((..(((((...........)))))...))))))))))................. (-21.82 = -23.61 + 1.80)
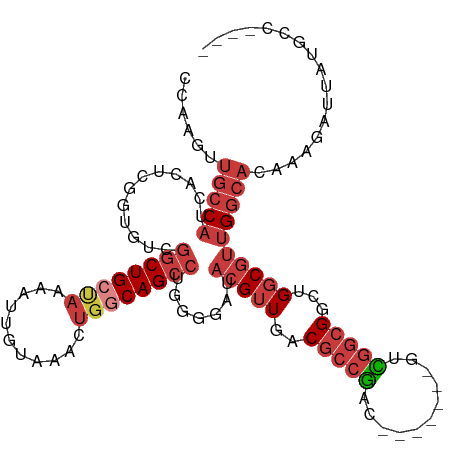
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:45:37 2011