| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,106,358 – 15,106,448 |
| Length | 90 |
| Max. P | 0.950655 |

| Location | 15,106,358 – 15,106,448 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 74.35 |
| Shannon entropy | 0.46246 |
| G+C content | 0.56477 |
| Mean single sequence MFE | -25.65 |
| Consensus MFE | -14.06 |
| Energy contribution | -16.78 |
| Covariance contribution | 2.72 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.580669 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
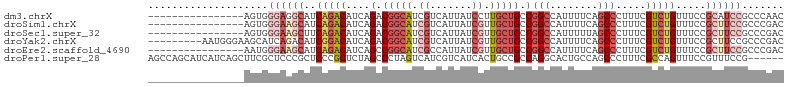
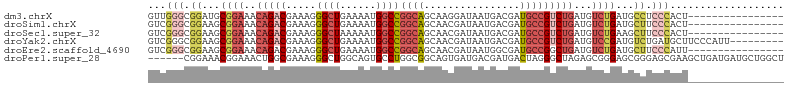
>dm3.chrX 15106358 90 + 22422827 ----------------AGUGGGAGGCAUCAGACAUCAGACGGCAUCGUCAUUAUCCUUGCUGCCGGCCAUUUUCAGCCCUUUCGUCUGUUUCCGCAUCCGCCCAAC ----------------..((((.((.(((.((...(((((((((.((..........)).))))(((........))).....)))))..)).).)))).)))).. ( -25.70, z-score = -1.34, R) >droSim1.chrX 11619794 90 + 17042790 ----------------AGUGGGAAGCAUCAGACAUCAGACGGCAUCGUCAUUAUCGUUGCUGCCGGCCAUUUUCAGCCCUUUCGUCUGUUUCCGCUUCCGCCCGAC ----------------....((((((..(((((....(.(((((.((.......)).))))).)(((........))).....))))).....))))))....... ( -27.70, z-score = -1.80, R) >droSec1.super_32 552829 90 + 562834 ----------------AGUGGGAAGCUUCAGACAUCAGACGGCAUCGUCAUUAUCGUUGCUGCCGGCCAUUUUUAGCCCUUUCGUCUGUUUCCGCUUCCGCCCGAC ----------------....((((((..(((((....(.(((((.((.......)).))))).)(((........))).....))))).....))))))....... ( -27.70, z-score = -1.92, R) >droYak2.chrX 9307412 97 + 21770863 ---------AAUGGGAAGCAUCAGACAUCGGACAUCAGACGGCAUCGUCAUUAUCGUUGCUGCCGGCCAUUUUCAGCCCUUUCGUCUGUUUCCGCUUCCGCCCGAC ---------....((((((....(....)(((...(((((((((.((.(......).)).))))(((........))).....)))))..)))))))))....... ( -30.50, z-score = -1.88, R) >droEre2.scaffold_4690 6841128 90 - 18748788 ----------------AAUGGGAAGCAUCAGACAUCAGCCGGCAUCGCCAUUAUCGUUGCUGCCGGCCAUUUUCAGCCCUUUCGUCUGUUUCCGCUUCCGCCCGAC ----------------....((((((..(((((....(((((((.((.(......).)).)))))))........(......)))))).....))))))....... ( -28.20, z-score = -1.92, R) >droPer1.super_28 907362 100 - 1111753 AGCCAGCAUCAUCAGCUUCGCUCCCGCUCCCGCUCUAGCCCUAGUCAUCGUCAUCACUGCCGCCAGGCACUGCCAGCCCUUUCGCCAGUUUCCGUUUCCG------ .((..(((......(((..(((...((....))...)))...)))............((((....)))).)))..)).......................------ ( -14.10, z-score = 0.86, R) >consensus ________________AGUGGGAAGCAUCAGACAUCAGACGGCAUCGUCAUUAUCGUUGCUGCCGGCCAUUUUCAGCCCUUUCGUCUGUUUCCGCUUCCGCCCGAC ....................((((((..(((((....(.(((((.((.......)).))))).)(((........))).....))))).....))))))....... (-14.06 = -16.78 + 2.72)
| Location | 15,106,358 – 15,106,448 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 74.35 |
| Shannon entropy | 0.46246 |
| G+C content | 0.56477 |
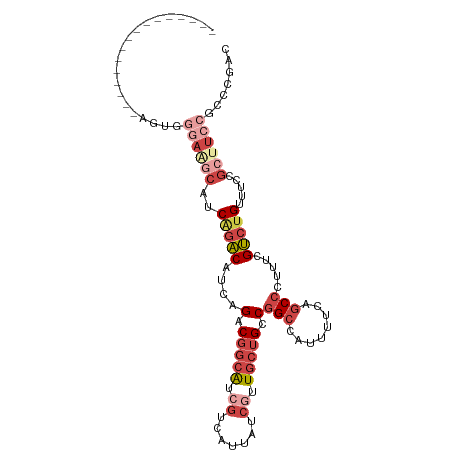
| Mean single sequence MFE | -32.75 |
| Consensus MFE | -18.80 |
| Energy contribution | -20.89 |
| Covariance contribution | 2.09 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.950655 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
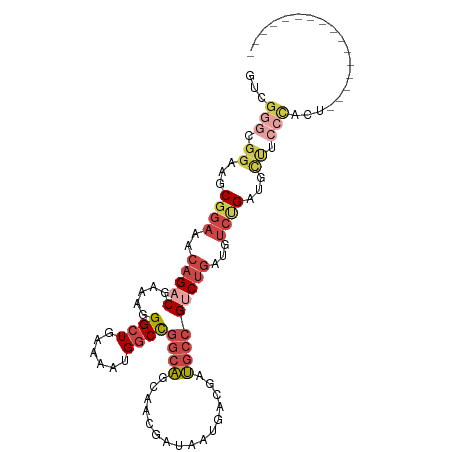
>dm3.chrX 15106358 90 - 22422827 GUUGGGCGGAUGCGGAAACAGACGAAAGGGCUGAAAAUGGCCGGCAGCAAGGAUAAUGACGAUGCCGUCUGAUGUCUGAUGCCUCCCACU---------------- ..((((.((...((((..(((((.....((((......))))((((....(........)..)))))))))...))))...)).))))..---------------- ( -34.50, z-score = -2.81, R) >droSim1.chrX 11619794 90 - 17042790 GUCGGGCGGAAGCGGAAACAGACGAAAGGGCUGAAAAUGGCCGGCAGCAACGAUAAUGACGAUGCCGUCUGAUGUCUGAUGCUUCCCACU---------------- ...(((.((...((((..(((((.....((((......))))((((.((.......))....)))))))))...))))...)).)))...---------------- ( -31.30, z-score = -1.85, R) >droSec1.super_32 552829 90 - 562834 GUCGGGCGGAAGCGGAAACAGACGAAAGGGCUAAAAAUGGCCGGCAGCAACGAUAAUGACGAUGCCGUCUGAUGUCUGAAGCUUCCCACU---------------- ...(((.((...((((..(((((.....(((((....)))))((((.((.......))....)))))))))...))))...)).)))...---------------- ( -31.70, z-score = -2.25, R) >droYak2.chrX 9307412 97 - 21770863 GUCGGGCGGAAGCGGAAACAGACGAAAGGGCUGAAAAUGGCCGGCAGCAACGAUAAUGACGAUGCCGUCUGAUGUCCGAUGUCUGAUGCUUCCCAUU--------- ((((((((....((((..(((((.....((((......))))((((.((.......))....)))))))))...)))).))))))))..........--------- ( -34.80, z-score = -1.96, R) >droEre2.scaffold_4690 6841128 90 + 18748788 GUCGGGCGGAAGCGGAAACAGACGAAAGGGCUGAAAAUGGCCGGCAGCAACGAUAAUGGCGAUGCCGGCUGAUGUCUGAUGCUUCCCAUU---------------- ...(((.((...((((..(((.(....)..))).....((((((((((..........))..))))))))....))))...)).)))...---------------- ( -32.00, z-score = -1.52, R) >droPer1.super_28 907362 100 + 1111753 ------CGGAAACGGAAACUGGCGAAAGGGCUGGCAGUGCCUGGCGGCAGUGAUGACGAUGACUAGGGCUAGAGCGGGAGCGGGAGCGAAGCUGAUGAUGCUGGCU ------((....))........(....)(((..(((...(((((((.((....))....)).)))))(((...((....))...)))...........)))..))) ( -32.20, z-score = -1.35, R) >consensus GUCGGGCGGAAGCGGAAACAGACGAAAGGGCUGAAAAUGGCCGGCAGCAACGAUAAUGACGAUGCCGUCUGAUGUCUGAUGCUUCCCACU________________ ...(((.((...((((..(((((.....((((......))))((((................)))))))))...))))...)).)))................... (-18.80 = -20.89 + 2.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:45:33 2011