| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,072,576 – 15,072,717 |
| Length | 141 |
| Max. P | 0.936665 |

| Location | 15,072,576 – 15,072,717 |
|---|---|
| Length | 141 |
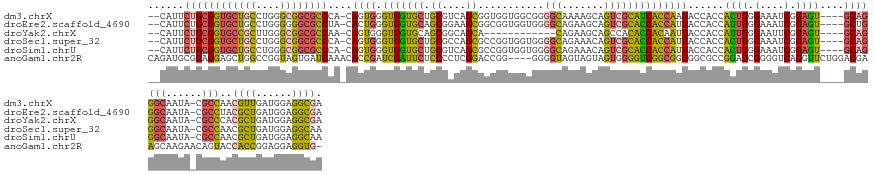
| Sequences | 6 |
| Columns | 149 |
| Reading direction | forward |
| Mean pairwise identity | 73.22 |
| Shannon entropy | 0.50000 |
| G+C content | 0.62820 |
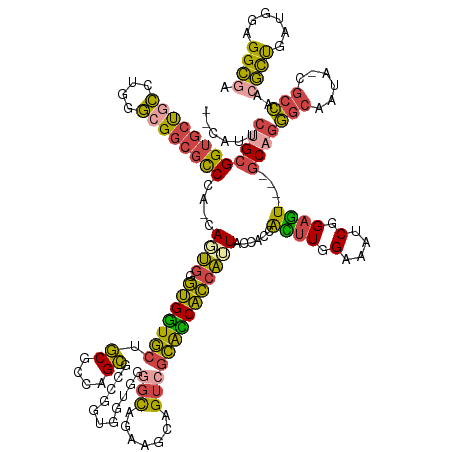
| Mean single sequence MFE | -62.67 |
| Consensus MFE | -36.71 |
| Energy contribution | -39.17 |
| Covariance contribution | 2.46 |
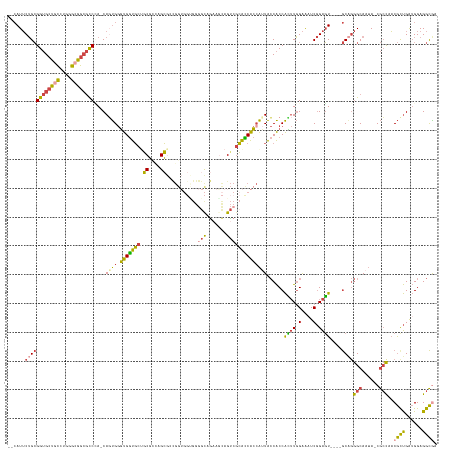
| Combinations/Pair | 1.36 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.936665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15072576 141 + 22422827 --CAUUCUGCGGUGCUGCCUGGGCGGCGCCCA-CAGUGGGUGGUGCUGCGUCAGCGGUGGUGGCGGGGCAAAAGCAGUCGCAUCACCAAUACCACCACUUGGAAAUCGGAGU----GCAGGGCAAUA-CGCCAACGUUGAUGGAGGCGA --..((((((((((((((....))))))))..-.....((((((((((...))))(((((((.(((.((....))..))))))))))...))))))((((.(....).))))----)))))).....-((((..((....))..)))). ( -63.20, z-score = -1.31, R) >droEre2.scaffold_4690 6808437 141 - 18748788 --CAUUCUGCGGUGCUGCCUGGGGGGCGCCUA-CACUGGGUGGUGCAGCGGAAGCGGCGGUGGUGGGGCAGAAGCAGUCGCACCACCAUUACCACCAUUUGGAAAUCGGAGU----GCUGGGCAAUA-CGCCUACGCUGAUGGAGGCGA --(((((((.((((((.(....).))))))..-((...((((((((.((....)).))(((((((.(((.......))).)))))))...))))))...)).....))))))----).(((((....-.)))))((((......)))). ( -66.90, z-score = -2.53, R) >droYak2.chrX 9262887 129 + 21770863 --CAUUCUGCGGUGCCGCUUGGGCGGCGCCAA-CAGUGGGUGGUGCAGCGGCAGCA------------CAGAAGCAGCCACACCACAAUUACCACCAUUUGGAAUUCGGAGU----GCAGGGCAAUA-CGCCCACGCUGAUGGAGGCGA --(((((((.((((((((....))))))))..-..(((((((((((....)).((.------------.....)).))))).)))).....(((.....)))....))))))----)..((((....-.)))).((((......)))). ( -55.40, z-score = -1.89, R) >droSec1.super_32 518819 141 + 562834 --CAUUCUGCGGUGCUGCCUGGGCGGCGCCCA-CAGUGGGUGGUGCUGCGCCAGCGCCGGUGGUGGGGCAGAAACAGUCGCACCACCAUUACCACCACUUGGAAAUCGGAGU----GCAGGGCAAUA-CGCCAACGCUGAUGGAGGCAA --.....(((((((((((....))))))))((-(((((((((((((((...)))))))(((((((.(((.......))).)))))))...)))..(((((.(....).))))----)...(((....-.)))..))))).))...))). ( -68.40, z-score = -1.89, R) >droSim1.chrU 4422300 141 - 15797150 --CAUUCUGCGGUGCUGCCUGGGCGGCGCCCA-CAGUGGGUGGUGCUGCGUCAGCGCCGGUGGUGGGGCAGAAACAGUCGCACCACCAUUACCACCACUUGGAAAUCGGAGU----GCAGGGCAAUA-CGCCAACGCUGAUGGAGGCAA --.....(((((((((((....))))))))((-(((((((((((((((...)))))))(((((((.(((.......))).)))))))...)))..(((((.(....).))))----)...(((....-.)))..))))).))...))). ( -68.40, z-score = -2.24, R) >anoGam1.chr2R 5362018 144 + 62725911 CAGAUGCGGAGGAGCUGGCCGGUAGUGAUCAAACACCGAUCGAUUCUCCCCUCGGACCGG----GGGGUAGUAGUAGUGGGGUCGGCGGUGGCGCCGGAUCGGGGUCAGGUUCUGGACGAAGCAAGAACAGUACCACCGGAGGAGGUG- ....(.(((.((..((((((....((......)).(((((((((((((((((((...)))----))))..........))))))((((....)))).))))))))))))(((((..........)))))....)).))).).......- ( -53.70, z-score = 0.22, R) >consensus __CAUUCUGCGGUGCUGCCUGGGCGGCGCCCA_CAGUGGGUGGUGCUGCGCCAGCGCCGGUGGUGGGGCAGAAGCAGUCGCACCACCAUUACCACCACUUGGAAAUCGGAGU____GCAGGGCAAUA_CGCCAACGCUGAUGGAGGCGA ......((((((((((((....))))))))........(((((((..((....))...(((((.(.(((.......))).).)))))..)))))))((((.(....).))))....))))(((......)))..((((......)))). (-36.71 = -39.17 + 2.46)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:45:27 2011