
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,053,177 – 15,053,281 |
| Length | 104 |
| Max. P | 0.793818 |

| Location | 15,053,177 – 15,053,281 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 81.24 |
| Shannon entropy | 0.32493 |
| G+C content | 0.42500 |
| Mean single sequence MFE | -27.23 |
| Consensus MFE | -14.48 |
| Energy contribution | -16.34 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.793818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
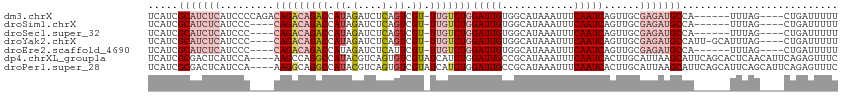
>dm3.chrX 15053177 104 - 22422827 UCAUCGCAUCUCAUCCCCAGACAGACAGACCAUAGAUCUCAGUCGU-UUGUCUGGAUUGUGGCAUAAAUUUCAAUCAGUUGCGAGAUGCCA------UUUAG----CUGAUUUUU .....(((((((....((((((((((.(((...........)))))-))))))))...(..((..............))..))))))))..------.....----......... ( -32.04, z-score = -3.43, R) >droSim1.chrX 11597760 100 - 17042790 UCAUCGCAUCUCAUCCC----CAGACAGACCAUAGAUCUCAGUCGU-UUGUCUGGAUUGUGGCAUAAAUUUCAAUCAGUUGCGAGAUGCCA------UUUAG----CUGAUUUUU .....(((((((....(----(((((((((.((.(....).)).))-))))))))...(..((..............))..))))))))..------.....----......... ( -30.14, z-score = -3.26, R) >droSec1.super_32 499899 100 - 562834 UCAUCGCAUCUCAUCCC----CAGACAGACCAUAGAUCUCAGUCGU-UUGUCUGGAUUGUGGCAUAAAUUUCAAUCAGUUGCGAGAUGCCA------UUUAG----CUGAUUUUU .....(((((((....(----(((((((((.((.(....).)).))-))))))))...(..((..............))..))))))))..------.....----......... ( -30.14, z-score = -3.26, R) >droYak2.chrX 9243010 105 - 21770863 UCAUCGCAUCUCAUCCC----CAGACAGACCAUAGAUCUCAGUCGU-UUGUCUGGAUUGUGGCAUAAAUUUCAAUCAGUUGCGAGAUGCCAUU-GCAUUUAG----CUGAUUUUU ..((((((((((....(----(((((((((.((.(....).)).))-))))))))...(..((..............))..))))))))....-((.....)----).))).... ( -31.14, z-score = -3.00, R) >droEre2.scaffold_4690 6788282 100 + 18748788 UCAUCGCAUCUCAUCCC----CAGACAGACCAUAGAUCUCAUUCGU-UUGUCUGGAUUGUGGCAUAAAUUUCAAUCAGUUGCGAGAUGCCA------UUUAG----CUGAUUUUU .....(((((((....(----(((((((((....((......))))-))))))))...(..((..............))..))))))))..------.....----......... ( -28.94, z-score = -3.07, R) >dp4.chrXL_group1a 7319776 111 + 9151740 UCAUCGCGACUCAUCCA----AAGCCAGGCCAUACGUCAGUGUCGUAUCAUCUGGAUUGCCGCAUAAAUUUCAAUCACUUGCAUUAAGCAUUCAGCACUCAACAUUCAGAGUUUC .......(((((.....----...(((((..(((((.......)))))..)))))......(((...............))).....((.....))............))))).. ( -18.06, z-score = -0.26, R) >droPer1.super_28 837805 111 + 1111753 UCAUCGCGACUCAUCCA----AAGGCAGGCCAUACGUCAGUGUCGUAUCAUCUGGAUUGCCGCAUAAAUUUCAAUCACUUGCAUUAAGCAUUCAGCAUUCAGCAUUCAGAGUUUC .......(((((.....----....((((..(((((.......)))))..))))((.(((.(((...............))).....((.....)).....))).)).))))).. ( -20.16, z-score = -0.36, R) >consensus UCAUCGCAUCUCAUCCC____CAGACAGACCAUAGAUCUCAGUCGU_UUGUCUGGAUUGUGGCAUAAAUUUCAAUCAGUUGCGAGAUGCCA______UUUAG____CUGAUUUUU .....(((((((.........(((((((((.((.(....).)).)).)))))))(((((............)))))......))))))).......................... (-14.48 = -16.34 + 1.86)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:45:26 2011