| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,424,798 – 11,424,859 |
| Length | 61 |
| Max. P | 0.554598 |

| Location | 11,424,798 – 11,424,859 |
|---|---|
| Length | 61 |
| Sequences | 10 |
| Columns | 70 |
| Reading direction | forward |
| Mean pairwise identity | 84.49 |
| Shannon entropy | 0.32352 |
| G+C content | 0.45276 |
| Mean single sequence MFE | -16.28 |
| Consensus MFE | -10.51 |
| Energy contribution | -10.94 |
| Covariance contribution | 0.43 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.554598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
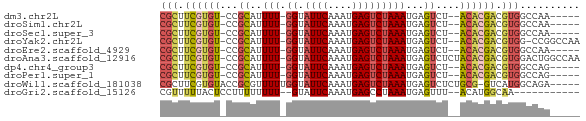
>dm3.chr2L 11424798 61 + 23011544 CGCUUCGUGU-CCGCAUUUU-GGUAUUCAAAUGAGUCUAAAUGAGUCU--ACACGACGUGGCCAA----- (((.((((((-..((..(((-((.((((....)))))))))...))..--)))))).))).....----- ( -17.20, z-score = -2.04, R) >droSim1.chr2L 11233898 61 + 22036055 CGCUUCGUGU-CCGCAUUUU-GGUAUUCAAAUGAGUCUAAAUGAGUCU--ACACGACGUGGCCAA----- (((.((((((-..((..(((-((.((((....)))))))))...))..--)))))).))).....----- ( -17.20, z-score = -2.04, R) >droSec1.super_3 6819717 61 + 7220098 CGCUUCGUGU-CCGCAUUUU-GGUAUUCAAAUGAGUCUAAAUGAGUCU--ACACGACGUGGCCAA----- (((.((((((-..((..(((-((.((((....)))))))))...))..--)))))).))).....----- ( -17.20, z-score = -2.04, R) >droYak2.chr2L 7824317 65 + 22324452 CGCUUCGUGU-CCGCAUUUU-GGUAUUCAAAUGAGUCUAAAUGAGUCU--ACACGACGUGG-CCGGCCAA (((.((((((-..((..(((-((.((((....)))))))))...))..--)))))).))).-........ ( -17.20, z-score = -0.95, R) >droEre2.scaffold_4929 12626530 61 - 26641161 CGCUUCGUGU-CCGCAUUUU-GGUAUUCAAAUGAGUCUAAAUGAGUCU--ACACGACGUGGCCAA----- (((.((((((-..((..(((-((.((((....)))))))))...))..--)))))).))).....----- ( -17.20, z-score = -2.04, R) >droAna3.scaffold_12916 12130339 68 - 16180835 CGCUUCGUGU-CCGCAUUUU-GGUAUUCAAAUGAGUCUAAAUGAGUCUCUACACGACGUGGACUGGCCAA .(((....((-((((..(((-((.((((....)))))))))...(((.......))))))))).)))... ( -19.60, z-score = -1.88, R) >dp4.chr4_group3 6878611 61 + 11692001 CGCUUCGUGU-CCGCAUUUU-GGUAUUCAAAUGAGUCUAAAUGAGUCU--ACACGACGUGGCCAG----- (((.((((((-..((..(((-((.((((....)))))))))...))..--)))))).))).....----- ( -17.20, z-score = -2.03, R) >droPer1.super_1 8326727 61 + 10282868 CGCUUCGUGU-CCGCAUUUU-GGUAUUCAAAUGAGUCUAAAUGAGUCU--ACACGACGUGGCCAG----- (((.((((((-..((..(((-((.((((....)))))))))...))..--)))))).))).....----- ( -17.20, z-score = -2.03, R) >droWil1.scaffold_181038 326335 64 - 637489 CGCUUCGUGUACCGCGUUUUUGGUAUUCAAAUGAGUCUAAAUGAGUCUCUGCG-GUCAUGGCAGA----- .((..((((.(((((...((((.....)))).(((.((.....)).))).)))-))))))))...----- ( -17.80, z-score = -1.58, R) >droGri2.scaffold_15126 7243111 55 - 8399593 CGUUUUUACUCCUUUUUUUU--GUAUUCAAAUGAGCCUAAAUGAGUUU--ACAUGGCAA----------- (((((.(((...........--)))...))))).(((...(((.....--.))))))..----------- ( -5.00, z-score = 0.36, R) >consensus CGCUUCGUGU_CCGCAUUUU_GGUAUUCAAAUGAGUCUAAAUGAGUCU__ACACGACGUGGCCAA_____ (((.((((((....(((((..((.((((....))))))))))).......)))))).))).......... (-10.51 = -10.94 + 0.43)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:32:35 2011