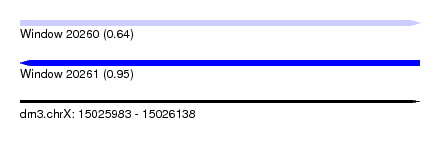
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,025,983 – 15,026,138 |
| Length | 155 |
| Max. P | 0.947715 |

| Location | 15,025,983 – 15,026,138 |
|---|---|
| Length | 155 |
| Sequences | 7 |
| Columns | 157 |
| Reading direction | forward |
| Mean pairwise identity | 78.77 |
| Shannon entropy | 0.40428 |
| G+C content | 0.51996 |
| Mean single sequence MFE | -50.82 |
| Consensus MFE | -24.86 |
| Energy contribution | -26.68 |
| Covariance contribution | 1.82 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.639183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
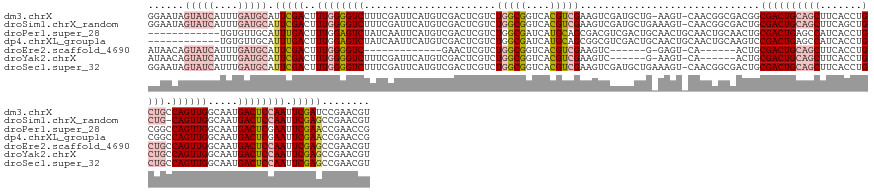
>dm3.chrX 15025983 155 + 22422827 ACGUUCGGAUCGAAUUGGAGUCAUUGCCAACUGGCAGCAGGUGAAGCUGCAGUCGCCGUCGCCGUUG-ACUU-CAGCAUCGACUUCGACGUGACCGCCAGACGAGUCGACAUGAAUCGAAAGACCCCAAAGUCGAAUGCAUCAAAUGAUACUAUUCC .(((((((.(((..((((((((.(((((....))))).....((.((((.(((((.((....)).))-))).-)))).))))))))))..))))))...))))..(((((.((..((....))...))..)))))....(((....)))........ ( -48.20, z-score = -0.88, R) >droSim1.chrX_random 3908894 155 + 5698898 ACGUUCGGCUCGAAUUGGAGUCAUUGCCAACUG-CAGCAGCUGAAGCUGCAGUCGCAGUCGCCGUUG-ACUUUCAGCAUCGACUUCGACGUGACCGCCAGACGAGUCGACAUGAAUCGAAAGACCCCAAAGUCGAAUGCAUCAAAUGAUACUAUUCC .(((((((((((..((((.((((((((.....)-))(((((....))))).((((.(((((..((((-.....))))..))))).)))))))))..)))).))))))(((.((..((....))...))..)))))))).(((....)))........ ( -53.50, z-score = -2.71, R) >droPer1.super_28 767890 145 - 1111753 CGGUUCGGUUCGAAUUCGAGUCAUUGCCAACUGGCCGCAGGUGAUGGCUCAGUCGCAGUUGCAGUUGCAGUUGCAGUCGACGUCGCUGCAUGAUCGCCAGACGAGUCGACAUGAAUUGAUAGACUCCAAAGUCAAAUGCAACACA------------ ....((((((((.....(((((((..((...........))..)))))))....(((..(((....)))..))).((((((.(((((((......).))).))))))))).))))))))..((((....))))............------------ ( -50.70, z-score = -1.11, R) >dp4.chrXL_group1a 7252645 145 - 9151740 CGGUUCGGUUCGAAUUCGAGUCAUUGCCAACUGGCCGCAGGUGAUGGCUCAGUCGGACUUGCAGUUGCAGUUGCAGUCGACGCCGCUGCAUGAUCGCCAGACGAGUCGACAUGAAUUGAUAGACUCCAAAGUCAAAUGCAACACA------------ ......(((((((....(((((((..((...........))..)))))))..)))))))(((....)))((((((((((((..((((((......).))).)).))))))...........((((....))))...))))))...------------ ( -51.80, z-score = -1.32, R) >droEre2.scaffold_4690 6761337 130 - 18748788 ACGUUCGGCUCGAAUUGGAGUCAUUGCCAACUGGCAGCAGGUGAAGCUGCAGUCGCAGU------UG-ACUC-C------GACUUCGACGUGACCGCCAGACGAGUUC-------------GACCCCAAAGUCGAAUGCAUCAAAUGAUACUGUUAU .(((..(((((((((((((((((((((..((((.((((.......)))))))).)))).------))-))))-)------)).))))).......)))..))).((((-------------(((......)))))))..(((....)))........ ( -48.71, z-score = -3.05, R) >droYak2.chrX 9216327 143 + 21770863 ACGUUCGGCUCGAAUUGGAGUCAUUGCCAACUGGCAGCAGGUGAAGCUGCAGUCGCAGU------UG-ACUU-C------GACUUCGACGUGACCGCCAGACGAGUCGACAUGAAUCGAAAGACCCCAAAGUCGAAUGCAUCAAAUGAUACUGUUAU .(((((((((((..((((.(((((((((....))))((((......)))).((((.(((------((-....-)------)))).)))))))))..)))).))))))(((.((..((....))...))..)))))))).(((....)))........ ( -48.50, z-score = -2.10, R) >droSec1.super_32 472802 156 + 562834 ACGUUCGGCUCGAAUUGGAGUCAUUGCCAACUGGCAGCAGGUGAAGCUGCAGUCGCAGUCGCCGUUG-ACUUUCAGCAUCGACUUCGACGUGACCGCCAGACGAGUCGACAUGAAUCGAAAGACCCCAAAGUCGAAUGCAUCAAAUGAUACUAUUCC .(((((((((((..((((.(((((((((....))))((((......)))).((((.(((((..((((-.....))))..))))).)))))))))..)))).))))))(((.((..((....))...))..)))))))).(((....)))........ ( -54.30, z-score = -2.40, R) >consensus ACGUUCGGCUCGAAUUGGAGUCAUUGCCAACUGGCAGCAGGUGAAGCUGCAGUCGCAGUCGCCGUUG_ACUU_CAGCAUCGACUUCGACGUGACCGCCAGACGAGUCGACAUGAAUCGAAAGACCCCAAAGUCGAAUGCAUCAAAUGAUACUAUUCC ....((((((((..((((........))))(((((.((((......)))).((((........((((......))))........))))......))))).))))))))............(((......)))........................ (-24.86 = -26.68 + 1.82)


| Location | 15,025,983 – 15,026,138 |
|---|---|
| Length | 155 |
| Sequences | 7 |
| Columns | 157 |
| Reading direction | reverse |
| Mean pairwise identity | 78.77 |
| Shannon entropy | 0.40428 |
| G+C content | 0.51996 |
| Mean single sequence MFE | -54.29 |
| Consensus MFE | -28.13 |
| Energy contribution | -30.16 |
| Covariance contribution | 2.03 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.947715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15025983 155 - 22422827 GGAAUAGUAUCAUUUGAUGCAUUCGACUUUGGGGUCUUUCGAUUCAUGUCGACUCGUCUGGCGGUCACGUCGAAGUCGAUGCUG-AAGU-CAACGGCGACGGCGACUGCAGCUUCACCUGCUGCCAGUUGGCAAUGACUCCAAUUCGAUCCGAACGU (((...(((((....)))))..((((..((((((((...........((((.(.(((.((((..((((((((....))))).))-).))-))))))))))(.((((((((((.......)))).)))))).)...)))))))).)))))))...... ( -62.20, z-score = -3.30, R) >droSim1.chrX_random 3908894 155 - 5698898 GGAAUAGUAUCAUUUGAUGCAUUCGACUUUGGGGUCUUUCGAUUCAUGUCGACUCGUCUGGCGGUCACGUCGAAGUCGAUGCUGAAAGU-CAACGGCGACUGCGACUGCAGCUUCAGCUGCUG-CAGUUGGCAAUGACUCCAAUUCGAGCCGAACGU ((....(((((....))))).(((((..((((((((..(((((....))))).(((((((((..((((((((....))))).)))..))-))..))))).((((((((((((.......))))-))))).)))..)))))))).)))))))...... ( -64.00, z-score = -3.65, R) >droPer1.super_28 767890 145 + 1111753 ------------UGUGUUGCAUUUGACUUUGGAGUCUAUCAAUUCAUGUCGACUCGUCUGGCGAUCAUGCAGCGACGUCGACUGCAACUGCAACUGCAACUGCGACUGAGCCAUCACCUGCGGCCAGUUGGCAAUGACUCGAAUUCGAACCGAACCG ------------.(.(((...(((((..((.(((((...........((((((((((...((......)).)))).))))))(((...(((....)))....((((((.(((.........))))))))))))..))))).)).)))))...)))). ( -43.90, z-score = -0.51, R) >dp4.chrXL_group1a 7252645 145 + 9151740 ------------UGUGUUGCAUUUGACUUUGGAGUCUAUCAAUUCAUGUCGACUCGUCUGGCGAUCAUGCAGCGGCGUCGACUGCAACUGCAACUGCAAGUCCGACUGAGCCAUCACCUGCGGCCAGUUGGCAAUGACUCGAAUUCGAACCGAACCG ------------.(.(((...(((((..((.(((((...........((((((((((...((......)).)))).))))))((((........)))).(.(((((((.(((.........)))))))))))...))))).)).)))))...)))). ( -43.70, z-score = 0.06, R) >droEre2.scaffold_4690 6761337 130 + 18748788 AUAACAGUAUCAUUUGAUGCAUUCGACUUUGGGGUC-------------GAACUCGUCUGGCGGUCACGUCGAAGUC------G-GAGU-CA------ACUGCGACUGCAGCUUCACCUGCUGCCAGUUGGCAAUGACUCCAAUUCGAGCCGAACGU ......(((((....))))).(((((((....))))-------------)))..(((.(((((....).(((((...------(-((((-((------..((((((((((((.......)))).))))).))).)))))))..))))))))).))). ( -52.20, z-score = -3.53, R) >droYak2.chrX 9216327 143 - 21770863 AUAACAGUAUCAUUUGAUGCAUUCGACUUUGGGGUCUUUCGAUUCAUGUCGACUCGUCUGGCGGUCACGUCGAAGUC------G-AAGU-CA------ACUGCGACUGCAGCUUCACCUGCUGCCAGUUGGCAAUGACUCCAAUUCGAGCCGAACGU ......(((((....))))).(((((..((((((((.((((((((((((.(((.((.....))))))))).).))))------)-))((-((------((((.....(((((.......)))))))))))))...)))))))).)))))........ ( -53.80, z-score = -3.30, R) >droSec1.super_32 472802 156 - 562834 GGAAUAGUAUCAUUUGAUGCAUUCGACUUUGGGGUCUUUCGAUUCAUGUCGACUCGUCUGGCGGUCACGUCGAAGUCGAUGCUGAAAGU-CAACGGCGACUGCGACUGCAGCUUCACCUGCUGCCAGUUGGCAAUGACUCCAAUUCGAGCCGAACGU ((....(((((....))))).(((((..((((((((..(((((....))))).(((((((((..((((((((....))))).)))..))-))..))))).((((((((((((.......)))).))))).)))..)))))))).)))))))...... ( -60.20, z-score = -2.83, R) >consensus GGAAUAGUAUCAUUUGAUGCAUUCGACUUUGGGGUCUUUCGAUUCAUGUCGACUCGUCUGGCGGUCACGUCGAAGUCGAUGCUG_AAGU_CAACGGCGACUGCGACUGCAGCUUCACCUGCUGCCAGUUGGCAAUGACUCCAAUUCGAGCCGAACGU ......(((((....))))).(((((..((((((((......................(((((....)))))..............................((((((((((.......)))).)))))).....)))))))).)))))........ (-28.13 = -30.16 + 2.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:45:24 2011