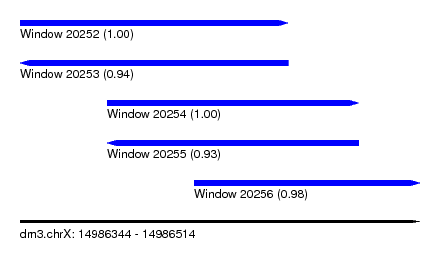
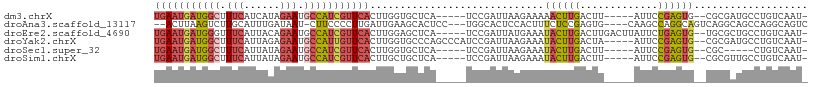
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,986,344 – 14,986,514 |
| Length | 170 |
| Max. P | 0.999598 |

| Location | 14,986,344 – 14,986,458 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.42 |
| Shannon entropy | 0.59562 |
| G+C content | 0.38947 |
| Mean single sequence MFE | -29.36 |
| Consensus MFE | -14.79 |
| Energy contribution | -17.68 |
| Covariance contribution | 2.89 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.06 |
| SVM RNA-class probability | 0.999598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14986344 114 + 22422827 GAGUCUCACACCAAAAUAAAGGACCUUCAUUAAAAGA---AAGAGGUUCAGGCAUCCUAUAAC---UUAAAGGCACUCAGUGAAUGAUGGCUUUCAUCAUAGAAUGCCAUCGUUCACUUG ((((.((.............(((((((..((....))---..)))))))(((...))).....---.....)).))))(((((((((((((.(((......))).))))))))))))).. ( -36.20, z-score = -5.00, R) >droAna3.scaffold_13117 1228168 89 + 5790199 GCUCCUCCUGGCACUUUCCCCGACCGACGCUAAGAGG--------AUUACAAAAAUACACA-----CUUAAGUCUUGCAUUUGAUAAUCUUCCCCUUGAUUG------------------ ..(((((.((((................)))).))))--------)...............-----.....(((........)))((((........)))).------------------ ( -12.19, z-score = -0.63, R) >droEre2.scaffold_4690 6722667 117 - 18748788 GAGUCUUACACAAAAAUAAACGACCUACAUUAAAAAAUAAAGGAGGUUCAGGAAUCUUAUGAC---UUCAAUGCAUUCCGUGAAUGAUGGGUUUCAUUACAGAAUGCCAUCGUUCACUUG .....................(((((.(.(((.....))).).)))))..((((((...((..---..))..).)))))(((((((((((..(((......)))..)))))))))))... ( -26.50, z-score = -2.04, R) >droYak2.chrX 9177469 119 + 21770863 GAGUCUUACACAAAAAUAACAGACCUACAUUAAGAAA-AAAGGAGGUUCAGAAAGCGUAUAAUAAAUUGAGUGCACUCAGUGAAUGAUGGCUUUCAUUAGAGAAUGCCAUUGUUCACUUG ((((.................(((((...((......-.))..)))))......(((..(((....)))..)))))))((((((..(((((.(((......))).)))))..)))))).. ( -28.80, z-score = -2.53, R) >droSec1.super_32 433086 114 + 562834 GAGUCUAACACCGAAAUAAAGGACCUUCAUUAAAAGA---AAGAGGUUCUGGGAUCCUAUAAC---UUAAAGGCACUCAGUGAAUGAUGGCUUUCAUUAUAGAAUGCCAUCGUUCACUUG ((((((.............((((((((..((....))---..))))))))((....)).....---.....)).))))(((((((((((((.(((......))).))))))))))))).. ( -36.00, z-score = -4.31, R) >droSim1.chrX 11563278 114 + 17042790 GAGUCUCACACCGAAAUAAAGGACCUUCAUUAAAAGA---AAGAGGUUCUGGGAUCCUAUAAC---UUAAAGGCACUCAGUGAAUGAUGGCUUUCAUUAUAGAAUGCCAUCGUUCACUUG ((((.((............((((((((..((....))---..))))))))((....)).....---.....)).))))(((((((((((((.(((......))).))))))))))))).. ( -36.50, z-score = -4.24, R) >consensus GAGUCUCACACCAAAAUAAAGGACCUACAUUAAAAGA___AAGAGGUUCAGGAAUCCUAUAAC___UUAAAGGCACUCAGUGAAUGAUGGCUUUCAUUAUAGAAUGCCAUCGUUCACUUG .....................(((((.................)))))...............................((((((((((((.(((......))).))))))))))))... (-14.79 = -17.68 + 2.89)

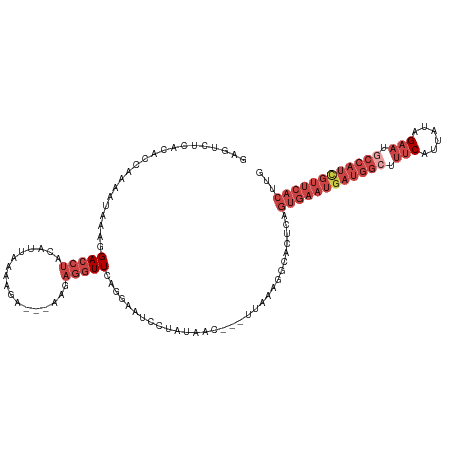
| Location | 14,986,344 – 14,986,458 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.42 |
| Shannon entropy | 0.59562 |
| G+C content | 0.38947 |
| Mean single sequence MFE | -28.04 |
| Consensus MFE | -11.27 |
| Energy contribution | -14.30 |
| Covariance contribution | 3.03 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.939498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14986344 114 - 22422827 CAAGUGAACGAUGGCAUUCUAUGAUGAAAGCCAUCAUUCACUGAGUGCCUUUAA---GUUAUAGGAUGCCUGAACCUCUU---UCUUUUAAUGAAGGUCCUUUAUUUUGGUGUGAGACUC ..((((((.((((((.(((......))).)))))).))))))((((........---(((.((((...)))))))(((..---.((...(((((((...)))))))..))...))))))) ( -31.90, z-score = -2.42, R) >droAna3.scaffold_13117 1228168 89 - 5790199 ------------------CAAUCAAGGGGAAGAUUAUCAAAUGCAAGACUUAAG-----UGUGUAUUUUUGUAAU--------CCUCUUAGCGUCGGUCGGGGAAAGUGCCAGGAGGAGC ------------------.((((........))))...(((((((..((....)-----).)))))))......(--------((((((.(((.(..((...))..)))).))))))).. ( -18.00, z-score = 0.18, R) >droEre2.scaffold_4690 6722667 117 + 18748788 CAAGUGAACGAUGGCAUUCUGUAAUGAAACCCAUCAUUCACGGAAUGCAUUGAA---GUCAUAAGAUUCCUGAACCUCCUUUAUUUUUUAAUGUAGGUCGUUUAUUUUUGUGUAAGACUC ..(((...(((((.(((((((((((((......))))).)))))))))))))..---..(((((((....(((((..(((.((((....)))).)))..))))).)))))))....))). ( -29.80, z-score = -2.57, R) >droYak2.chrX 9177469 119 - 21770863 CAAGUGAACAAUGGCAUUCUCUAAUGAAAGCCAUCAUUCACUGAGUGCACUCAAUUUAUUAUACGCUUUCUGAACCUCCUUU-UUUCUUAAUGUAGGUCUGUUAUUUUUGUGUAAGACUC ..((((((..(((((.(((......))).)))))..))))))(((....))).........(((((......(((..(((..-...........)))...)))......)))))...... ( -26.62, z-score = -2.29, R) >droSec1.super_32 433086 114 - 562834 CAAGUGAACGAUGGCAUUCUAUAAUGAAAGCCAUCAUUCACUGAGUGCCUUUAA---GUUAUAGGAUCCCAGAACCUCUU---UCUUUUAAUGAAGGUCCUUUAUUUCGGUGUUAGACUC ..((((((.((((((.(((......))).)))))).))))))(((..(((((..---((((.((((....((.....)).---)))).)))))))))..))).................. ( -30.90, z-score = -2.62, R) >droSim1.chrX 11563278 114 - 17042790 CAAGUGAACGAUGGCAUUCUAUAAUGAAAGCCAUCAUUCACUGAGUGCCUUUAA---GUUAUAGGAUCCCAGAACCUCUU---UCUUUUAAUGAAGGUCCUUUAUUUCGGUGUGAGACUC ..((((((.((((((.(((......))).)))))).))))))(((..(((((..---((((.((((....((.....)).---)))).)))))))))..)))..((((.....))))... ( -31.00, z-score = -2.31, R) >consensus CAAGUGAACGAUGGCAUUCUAUAAUGAAAGCCAUCAUUCACUGAGUGCCUUUAA___GUUAUAGGAUUCCUGAACCUCUU___UCUUUUAAUGAAGGUCCUUUAUUUUGGUGUGAGACUC ..((((((.((((((.(((......))).)))))).)))))).........................................((((....(((((........)))))....))))... (-11.27 = -14.30 + 3.03)

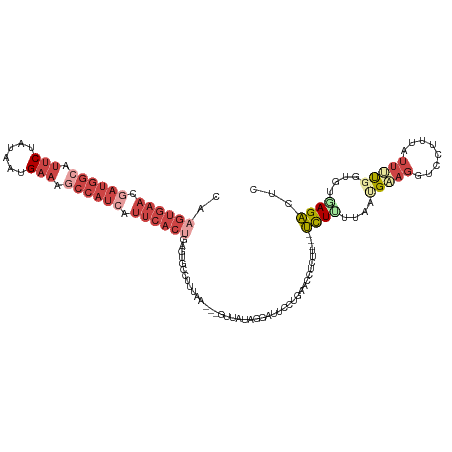
| Location | 14,986,381 – 14,986,488 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.29 |
| Shannon entropy | 0.24777 |
| G+C content | 0.39948 |
| Mean single sequence MFE | -35.61 |
| Consensus MFE | -24.10 |
| Energy contribution | -23.94 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.79 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.15 |
| SVM RNA-class probability | 0.997674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14986381 107 + 22422827 AAGAGGUUCAGGCAUCCUAUAAC---UUAAAGGCACUCAGUGAAUGAUGGCUUUCAUCAUAGAAUGCCAUCGUUCACUUGG-----UGCUCAUCCGAUUAAGAAAAACUUGACUU----- ..(((.((((((...))).....---.....((((((.(((((((((((((.(((......))).))))))))))))).))-----))))...........)))...))).....----- ( -36.70, z-score = -4.55, R) >droEre2.scaffold_4690 6722707 112 - 18748788 AGGAGGUUCAGGAAUCUUAUGAC---UUCAAUGCAUUCCGUGAAUGAUGGGUUUCAUUACAGAAUGCCAUCGUUCACUUGG-----AGCUCAUCCGAUUAUGAAAUACUUGACUUGACUU ..(((.((((..((((..(((((---(((((........(((((((((((..(((......)))..)))))))))))))))-----)).))))..)))).))))...))).......... ( -32.60, z-score = -2.12, R) >droYak2.chrX 9177508 115 + 21770863 AGGAGGUUCAGAAAGCGUAUAAUAAAUUGAGUGCACUCAGUGAAUGAUGGCUUUCAUUAGAGAAUGCCAUUGUUCACUUGGUGCCCAGCCCAUCCGAUUAAGAAAUACUUGACUA----- .(((((.((((...............))))(.(((((.((((((..(((((.(((......))).)))))..)))))).))))).)...)).)))..(((((.....)))))...----- ( -34.96, z-score = -3.05, R) >droSec1.super_32 433123 107 + 562834 AAGAGGUUCUGGGAUCCUAUAAC---UUAAAGGCACUCAGUGAAUGAUGGCUUUCAUUAUAGAAUGCCAUCGUUCACUUGG-----UGCUCAUCCGAUUAAGAAAUACUUGACUU----- ...(((((.(((....))).)))---))...((((((.(((((((((((((.(((......))).))))))))))))).))-----)))).......(((((.....)))))...----- ( -39.40, z-score = -5.27, R) >droSim1.chrX 11563315 107 + 17042790 AAGAGGUUCUGGGAUCCUAUAAC---UUAAAGGCACUCAGUGAAUGAUGGCUUUCAUUAUAGAAUGCCAUCGUUCACUUGC-----UGCUCAUCCGAUUAAGAAAUACUUGACUU----- ..((((((.(((....))).)))---.....((((...(((((((((((((.(((......))).))))))))))))))))-----).)))......(((((.....)))))...----- ( -34.40, z-score = -3.98, R) >consensus AAGAGGUUCAGGAAUCCUAUAAC___UUAAAGGCACUCAGUGAAUGAUGGCUUUCAUUAUAGAAUGCCAUCGUUCACUUGG_____UGCUCAUCCGAUUAAGAAAUACUUGACUU_____ ..(((.(((.(((..(((.(((....))).)))..))).((((((((((((.(((......))).))))))))))))........................)))...))).......... (-24.10 = -23.94 + -0.16)

| Location | 14,986,381 – 14,986,488 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.29 |
| Shannon entropy | 0.24777 |
| G+C content | 0.39948 |
| Mean single sequence MFE | -30.24 |
| Consensus MFE | -19.34 |
| Energy contribution | -20.06 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.934274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14986381 107 - 22422827 -----AAGUCAAGUUUUUCUUAAUCGGAUGAGCA-----CCAAGUGAACGAUGGCAUUCUAUGAUGAAAGCCAUCAUUCACUGAGUGCCUUUAA---GUUAUAGGAUGCCUGAACCUCUU -----...(((.((..((((((((..((.(.(((-----(..((((((.((((((.(((......))).)))))).))))))..))))).))..---)))).)))).)).)))....... ( -34.20, z-score = -3.70, R) >droEre2.scaffold_4690 6722707 112 + 18748788 AAGUCAAGUCAAGUAUUUCAUAAUCGGAUGAGCU-----CCAAGUGAACGAUGGCAUUCUGUAAUGAAACCCAUCAUUCACGGAAUGCAUUGAA---GUCAUAAGAUUCCUGAACCUCCU ...(((..(((((((((((......(((.....)-----))..(((((.(((((..(((......)))..))))).)))))))))))).))))(---(((....))))..)))....... ( -28.00, z-score = -1.86, R) >droYak2.chrX 9177508 115 - 21770863 -----UAGUCAAGUAUUUCUUAAUCGGAUGGGCUGGGCACCAAGUGAACAAUGGCAUUCUCUAAUGAAAGCCAUCAUUCACUGAGUGCACUCAAUUUAUUAUACGCUUUCUGAACCUCCU -----..................(((((.(((((((((((..((((((..(((((.(((......))).)))))..))))))..)))).).))...........)))))))))....... ( -30.40, z-score = -2.22, R) >droSec1.super_32 433123 107 - 562834 -----AAGUCAAGUAUUUCUUAAUCGGAUGAGCA-----CCAAGUGAACGAUGGCAUUCUAUAAUGAAAGCCAUCAUUCACUGAGUGCCUUUAA---GUUAUAGGAUCCCAGAACCUCUU -----.....(((...((((.....((((..(((-----(..((((((.((((((.(((......))).)))))).))))))..))))(((...---.....))))))).))))...))) ( -31.10, z-score = -3.57, R) >droSim1.chrX 11563315 107 - 17042790 -----AAGUCAAGUAUUUCUUAAUCGGAUGAGCA-----GCAAGUGAACGAUGGCAUUCUAUAAUGAAAGCCAUCAUUCACUGAGUGCCUUUAA---GUUAUAGGAUCCCAGAACCUCUU -----.....(((...((((.....((((..(((-----...((((((.((((((.(((......))).)))))).))))))...)))(((...---.....))))))).))))...))) ( -27.50, z-score = -2.08, R) >consensus _____AAGUCAAGUAUUUCUUAAUCGGAUGAGCA_____CCAAGUGAACGAUGGCAUUCUAUAAUGAAAGCCAUCAUUCACUGAGUGCCUUUAA___GUUAUAGGAUCCCUGAACCUCUU ............((((((((((......))))..........((((((.((((((.(((......))).)))))).))))))))))))................................ (-19.34 = -20.06 + 0.72)


| Location | 14,986,418 – 14,986,514 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 72.45 |
| Shannon entropy | 0.50392 |
| G+C content | 0.43781 |
| Mean single sequence MFE | -27.34 |
| Consensus MFE | -13.83 |
| Energy contribution | -14.78 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.978491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14986418 96 + 22422827 UGAAUGAUGGCUUUCAUCAUAGAAUGCCAUCGUUCACUUGGUGCUCA-----UCCGAUUAAGAAAAACUUGACUU-----AUUCCGAGUG--CGCGAUGCCUGUCAAU- (((((((((((.(((......))).)))))))))))....((((...-----((.((.((((..........)))-----).)).))..)--)))(((....)))...- ( -29.60, z-score = -3.09, R) >droAna3.scaffold_13117 1228220 99 + 5790199 --ACUUAAGUCUUGCAUUUGAUAAU-CUUCCCCUUGAUUGAAGCACUCC---UGGCACUCCACUUUCUCCGAGUG----CAAGCCAGGCAGUCAGGCAGCCAGGCAGUC --......((((.((......((((-(........)))))..(((((((---((((....(((((.....)))))----...)))))).)))...)).)).)))).... ( -28.20, z-score = -0.88, R) >droEre2.scaffold_4690 6722744 101 - 18748788 UGAAUGAUGGGUUUCAUUACAGAAUGCCAUCGUUCACUUGGAGCUCA-----UCCGAUUAUGAAAUACUUGACUUGACUUAUUCUGAGUG--UGCGCUGCCUGUCAAU- ((((((((((..(((......)))..))))))))))...((((((((-----(......)))).((((((...............)))))--)..))).)).......- ( -23.16, z-score = -0.43, R) >droYak2.chrX 9177548 101 + 21770863 UGAAUGAUGGCUUUCAUUAGAGAAUGCCAUUGUUCACUUGGUGCCCAGCCCAUCCGAUUAAGAAAUACUUGACUA-----AUUCCGAGUG--CGCGAUGCCUGUCAAU- ((((..(((((.(((......))).)))))..)))).........(((..((((((.........((((((....-----....))))))--)).)))).))).....- ( -24.80, z-score = -1.42, R) >droSec1.super_32 433160 91 + 562834 UGAAUGAUGGCUUUCAUUAUAGAAUGCCAUCGUUCACUUGGUGCUCA-----UCCGAUUAAGAAAUACUUGACUU-----AUUCCGAGUG--CGC-----CUGUCAAU- (((((((((((.(((......))).)))))))))))...(((((...-----((.((.((((..........)))-----).)).))..)--)))-----).......- ( -31.50, z-score = -4.84, R) >droSim1.chrX 11563352 96 + 17042790 UGAAUGAUGGCUUUCAUUAUAGAAUGCCAUCGUUCACUUGCUGCUCA-----UCCGAUUAAGAAAUACUUGACUU-----AUUCCGAGUG--CGCGUUGCCUGUCAAU- (((((((((((.(((......))).)))))))))))...((.((...-----((.((.((((..........)))-----).)).))..)--))).............- ( -26.80, z-score = -2.70, R) >consensus UGAAUGAUGGCUUUCAUUAUAGAAUGCCAUCGUUCACUUGGUGCUCA_____UCCGAUUAAGAAAUACUUGACUU_____AUUCCGAGUG__CGCGAUGCCUGUCAAU_ .((((((((((.(((......))).))))))))))((((((..........................................)))))).................... (-13.83 = -14.78 + 0.95)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:45:19 2011