
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,954,538 – 14,954,631 |
| Length | 93 |
| Max. P | 0.642818 |

| Location | 14,954,538 – 14,954,631 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 65.77 |
| Shannon entropy | 0.58371 |
| G+C content | 0.68361 |
| Mean single sequence MFE | -46.30 |
| Consensus MFE | -20.68 |
| Energy contribution | -19.83 |
| Covariance contribution | -0.86 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.642818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
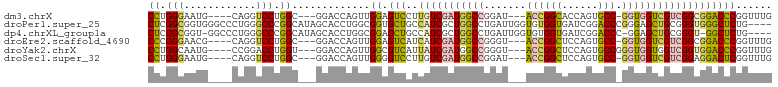
>dm3.chrX 14954538 93 - 22422827 CCUGGGAAUG----CAGGUCCUGGC---GGACCAGUUGGAGUCCUUGUCGAUGGCCGGAU---ACCGGCACCAGUGCC-GGUGGUCGUCGGCGGACCCGGUUUG .(((((....----..(((((....---)))))........(((..((((((((((....---(((((((....))))-))))))))))))))))))))).... ( -47.40, z-score = -2.41, R) >droPer1.super_25 1031724 100 + 1448063 CUCGGCGGUGGGCCCUGGGCCCGGCAUAGCACCUGGCGGUGCUGCCAUCGCUGGCCUGAUUGGUGUGGUGAUCGGACCCGGGAGCUGCGGGUGGGCUCUG---- ..((((((((((((...)))))((((..(((((....))))))))))))))))(((((.....((..((..(((....)))..))..))..)))))....---- ( -47.80, z-score = 0.17, R) >dp4.chrXL_group1a 6875097 97 - 9151740 CUCGGCGGU-GGCCCUGGGCCCGGCAUAGCACCUGGCGGAGCUGCCAUCGCUGGCCUGAUUGGUGUGGUGAUCGGACCC-GGAGCUGCGGGU-GGCUCUG---- .(((.(.(.-.((.(((((.(((((((.(((((...(((.(((((....)).))))))...))))).))).)))).)))-)).))..).).)-)).....---- ( -45.00, z-score = 0.17, R) >droEre2.scaffold_4690 6690230 93 + 18748788 CCCGGGAACG----CAGGUCCUGGC---GGACCAGUUGGAGUCAUCAUCGAUGGCCGGGU---ACCGGCUCCAGUGCC-GGUGGUCGUCGGCGGACCCGGUUUG .(((((..((----(.(((((....---)))))...............((((((((....---((((((......)))-))))))))))))))..))))).... ( -48.90, z-score = -2.35, R) >droYak2.chrX 9146882 94 - 21770863 CCUGGCAAUG----CCGGACCUGGU---GGACCAGUUGGCGUCAUUAUCGAUGGCCGGGU---ACCGGCUCCAGUGCCGGGUGGUGGUCGGUGGACCCGGUUUG (((((((..(----(((((((((((---...((....))((((......)))))))))))---.))))).....)))))))..(.((((....)))))...... ( -43.70, z-score = -1.03, R) >droSec1.super_32 401786 93 - 562834 CCUGGGAAUG----CAGGUCCUGGC---GGACCAGUUGGGGUCCUUGUCGAUGGCCGGAU---ACCGGCUCCAGUGCC-GGUGGUCGUCGGAGGACUCGGUUUG ((........----..(((((....---))))).....((((((((..((((((((....---((((((......)))-))))))))))))))))))))).... ( -45.00, z-score = -2.00, R) >consensus CCCGGCAAUG____CAGGUCCUGGC___GGACCAGUUGGAGUCAUCAUCGAUGGCCGGAU___ACCGGCUCCAGUGCC_GGUGGUCGUCGGUGGACCCGGUUUG ((.(((............))).)).............((.(((..(((((((((((.......((((((......))).)))))))))))))))))))...... (-20.68 = -19.83 + -0.86)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:45:10 2011