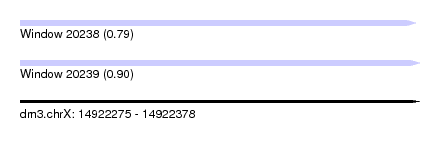
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,922,275 – 14,922,378 |
| Length | 103 |
| Max. P | 0.895016 |

| Location | 14,922,275 – 14,922,377 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 66.44 |
| Shannon entropy | 0.61334 |
| G+C content | 0.44354 |
| Mean single sequence MFE | -26.40 |
| Consensus MFE | -11.15 |
| Energy contribution | -11.43 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.787807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
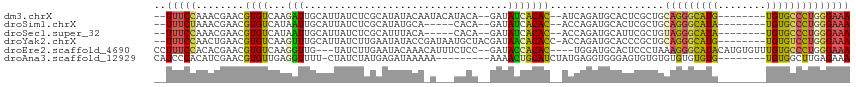
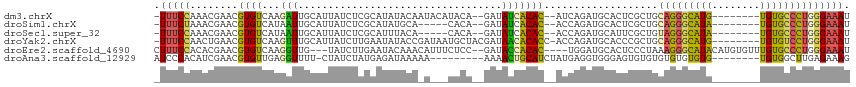
>dm3.chrX 14922275 102 + 22422827 --UUUCCAAACGAACGUGUCAAGAUUGCAUUAUCUCGCAUAUACAAUACAUACA--GAUAUCACAC--AUCAGAUGCACUCGCUGCAGGGCAUG--------UGUGCCCUGGGAAA --.((((...(((..(((((.....(((........)))...............--(((.......--))).)))))..)))...(((((((..--------..))))))))))). ( -25.30, z-score = -0.95, R) >droSim1.chrX 11508995 97 + 17042790 --UUUCUAAACGAACGUGUCAUAAUUGCAUUAUCUCGCAUAUGCA-----CACA--GAUAUCACAC--ACCAGAUGCACUCGCUGCAGGGCAUA--------UGUGCCCUGGGAAA --.((((...(((..((((((((..(((........))))))).)-----))).--(.((((....--....)))))..)))...(((((((..--------..))))))))))). ( -26.40, z-score = -1.65, R) >droSec1.super_32 377056 97 + 562834 --UUUCCAAACGAACGUGUCAUAAUUGCAUUAUCUCGCAUUUACA-----CACA--GAUAUCACAC--ACCAGAUGCAUUCGCUGUAGGGCAUA--------UGUGCCCUGGGAAA --.((((...((((.((((..(((.(((........))).))).)-----))).--(.((((....--....))))).))))...(((((((..--------..))))))))))). ( -25.40, z-score = -1.91, R) >droYak2.chrX 9123156 105 + 21770863 --UUUCCAACUGAACGUGUCAAGUUUGCAUUAUCUUGAAUAUACCGAUAAUGCUACGAUAACACACC-ACCAGAUGCACCCGCUGCAGGGCAUG--------UGUGUCCUGGGAAA --.((((........((((...((..((((((((...........)))))))).))....))))...-..(((((((((..(((....)))..)--------))))).))))))). ( -28.50, z-score = -1.48, R) >droEre2.scaffold_4690 6666922 107 - 18748788 CCUUUCCACACGAACGUGUCAAGGUUG---UAUCUUGAAUACAAACAUUUCUCC--GAUACCACAC----UGGAUGCACUCCCUAAAGGGCAUACAUGUGUUUGUGCCCUGGGAAA ...............(((((.((((.(---((((..(((.........)))...--))))).)).)----).)))))..((((...((((((((........)))))))))))).. ( -29.90, z-score = -1.66, R) >droAna3.scaffold_12929 43399 98 + 3277472 CAUCCCACAUCGAACGUGUUGAGGUUUU-CUAUCUAUGAGAUAAAAA---------AAAACUGCAUCUAUGAGGUGGGAGUGUGUGUGUGUGUG--------UGUGGCUUGAGAAA ((..((((((...(((..(......(((-((((((...((((.....---------........))))...))))))))).....)..)))..)--------)))))..))..... ( -22.92, z-score = -1.02, R) >consensus __UUUCCAAACGAACGUGUCAAGAUUGCAUUAUCUCGCAUAUACA_A___CACA__GAUAUCACAC__ACCAGAUGCACUCGCUGCAGGGCAUA________UGUGCCCUGGGAAA ...((((..................((((((.........................................)))))).......(((((((((........))))))))))))). (-11.15 = -11.43 + 0.28)
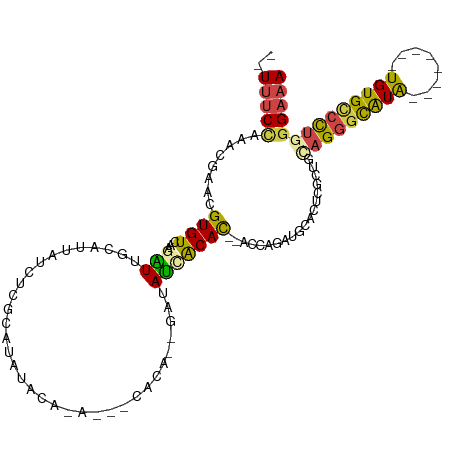
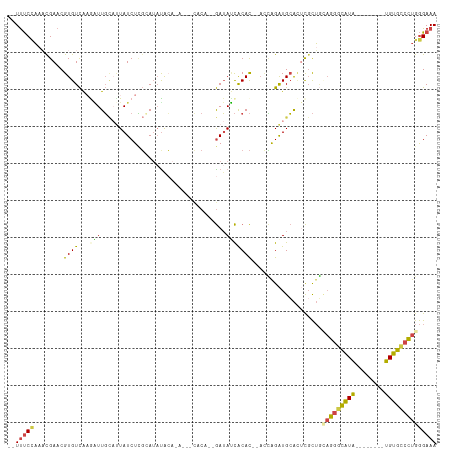
| Location | 14,922,275 – 14,922,378 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 66.75 |
| Shannon entropy | 0.61103 |
| G+C content | 0.43904 |
| Mean single sequence MFE | -26.45 |
| Consensus MFE | -11.76 |
| Energy contribution | -12.05 |
| Covariance contribution | 0.29 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.895016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14922275 103 + 22422827 -UUUCCAAACGAACGUGUCAAGAUUGCAUUAUCUCGCAUAUACAAUACAUACA--GAUAUCACAC--AUCAGAUGCACUCGCUGCAGGGCAUG--------UGUGCCCUGGGAAAU -(((((...(((..(((((.....(((........)))...............--(((.......--))).)))))..)))...(((((((..--------..)))))))))))). ( -25.80, z-score = -1.19, R) >droSim1.chrX 11508995 98 + 17042790 -UUUCUAAACGAACGUGUCAUAAUUGCAUUAUCUCGCAUAUGCA-----CACA--GAUAUCACAC--ACCAGAUGCACUCGCUGCAGGGCAUA--------UGUGCCCUGGGAAAU -(((((...(((..((((((((..(((........))))))).)-----))).--(.((((....--....)))))..)))...(((((((..--------..)))))))))))). ( -26.90, z-score = -1.89, R) >droSec1.super_32 377056 98 + 562834 -UUUCCAAACGAACGUGUCAUAAUUGCAUUAUCUCGCAUUUACA-----CACA--GAUAUCACAC--ACCAGAUGCAUUCGCUGUAGGGCAUA--------UGUGCCCUGGGAAAU -(((((...((((.((((..(((.(((........))).))).)-----))).--(.((((....--....))))).))))...(((((((..--------..)))))))))))). ( -25.90, z-score = -2.17, R) >droYak2.chrX 9123156 106 + 21770863 -UUUCCAACUGAACGUGUCAAGUUUGCAUUAUCUUGAAUAUACCGAUAAUGCUACGAUAACACACC-ACCAGAUGCACCCGCUGCAGGGCAUG--------UGUGUCCUGGGAAAU -(((((........((((...((..((((((((...........)))))))).))....))))...-..(((((((((..(((....)))..)--------))))).)))))))). ( -29.00, z-score = -1.67, R) >droEre2.scaffold_4690 6666923 107 - 18748788 CUUUCCACACGAACGUGUCAAGGUUG---UAUCUUGAAUACAAACAUUUCUCC--GAUACCACAC----UGGAUGCACUCCCUAAAGGGCAUACAUGUGUUUGUGCCCUGGGAAAU ..............(((((.((((.(---((((..(((.........)))...--))))).)).)----).)))))..((((...((((((((........))))))))))))... ( -29.90, z-score = -1.80, R) >droAna3.scaffold_12929 43400 98 + 3277472 AUCCCACAUCGAACGUGUUGAGGUUUU-CUAUCUAUGAGAUAAAAA---------AAAACUGCAUCUAUGAGGUGGGAGUGUGUGUGUGUGUG--------UGUGGCUUGAGAAAG ...((((((...(((..(......(((-((((((...((((.....---------........))))...))))))))).....)..)))..)--------))))).......... ( -21.22, z-score = -0.64, R) >consensus _UUUCCAAACGAACGUGUCAAGAUUGCAUUAUCUCGCAUAUACA_A___CACA__GAUAUCACAC__ACCAGAUGCACUCGCUGCAGGGCAUA________UGUGCCCUGGGAAAU .(((((..................((((((.........................................)))))).......(((((((((........)))))))))))))). (-11.76 = -12.05 + 0.29)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:45:05 2011