| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,905,559 – 14,905,649 |
| Length | 90 |
| Max. P | 0.915672 |

| Location | 14,905,559 – 14,905,649 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 65.23 |
| Shannon entropy | 0.77333 |
| G+C content | 0.48976 |
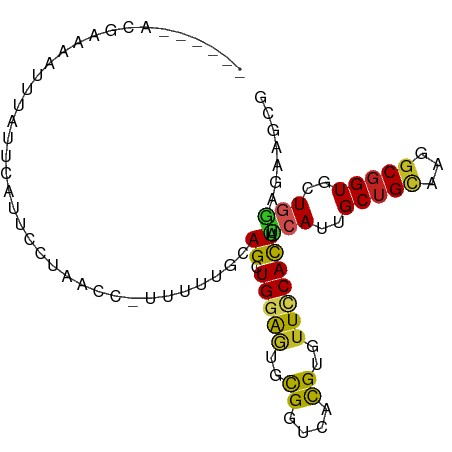
| Mean single sequence MFE | -25.64 |
| Consensus MFE | -11.80 |
| Energy contribution | -11.21 |
| Covariance contribution | -0.59 |
| Combinations/Pair | 1.53 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.915672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
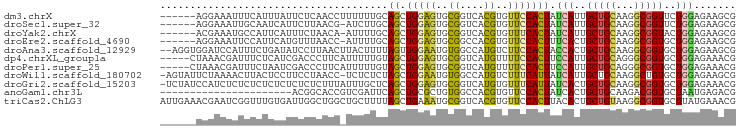
>dm3.chrX 14905559 90 - 22422827 ------AGGAAAUUUCAUUUAUUCUCAACCUUUUUUGCAGCUGGAGUGCGGUCACGUGUUCCACUAUCAUUGCUGCAAGGCGGUUCUGGAGAAGCG ------...............(((((((((..((((((((((((((..((....))..)))))........))))))))).))))...)))))... ( -29.10, z-score = -2.29, R) >droSec1.super_32 361494 89 - 562834 ------AGGAAAUUGCAAUCAUUCUUAACG-AUCUUGCAGCUGGAGUGCGGUCACGUGUUCCACUAUCAUUGCUGCAAGGCGGUUCUGGAGAAGCG ------........((..((.(((..(((.-.((((((((((((((..((....))..)))))........)))))))))..)))..))))).)). ( -29.60, z-score = -1.74, R) >droYak2.chrX 9107435 89 - 21770863 ------ACGAAAUGCCAUUCAUUUCUAACA-AUUUUGCAGCUGGAGUGCGGUCACGUGUUUCACUAUCAUUGCUGCAAGGCGGUACUGGAGAAGCG ------..(((......))).((((((((.-.((((((((((((((..((....))..)))))........)))))))))..))..)))))).... ( -23.90, z-score = -0.45, R) >droEre2.scaffold_4690 6651062 89 + 18748788 ------AGGAAAUUCCAUUCAUGUUUAACC-AUUUUGCAGCUGGAGUGCGGCCACGUGUUCCACUUUCACUGCUGCAAGGCGGUGCUGGAGAAGCG ------.((.....))...........(((-.((((((((((((((..((....))..)))))........))))))))).)))(((.....))). ( -30.60, z-score = -1.39, R) >droAna3.scaffold_12929 16085 94 - 3277472 --AGGUGGAUCCAUUUCUGAUAUCCUUAACUUACUUUUAGUUGGAAUGUGGCCAUGUCUUCCACUACCACUGCUGCAAGGCGGUGCUGGAGAAGCG --..((((..(((((((..((..................))..))).))))))))(.((((..(((.(((((((....))))))).))).))))). ( -24.87, z-score = -0.19, R) >dp4.chrXL_group1a 6833990 91 - 9151740 -----CUAAACGAUUUCUCAUCGACCCUUCAUUUUUGUAGCUGGAGUGCGGUCAUGUUUUCCACUUCCAUUGCUGCAGGGCGGUGCUGGAGAAACG -----.....((.((((((((((.((((........((((((((((((.((........))))).))))..)))))))))))))....)))))))) ( -26.90, z-score = -1.30, R) >droPer1.super_25 987882 91 + 1448063 -----CUAAACGAUUUCUAAUCGACCCUUCAUUUUUGUAGCUGGAGUGCGGUCAUGUUUUCCACUUCCAUUGCUGCAGGGCGGUGCUGGAGAAACG -----.....((.(((((.((((.((((........((((((((((((.((........))))).))))..))))))))))))).....))))))) ( -25.40, z-score = -1.00, R) >droWil1.scaffold_180702 4140662 94 - 4511350 -AGUAUUCUAAAACUUACUCCUUCCUAACC-UCUCUCUAGCUGGAAUGUGGCCAUGUCUUUCAUUAUCAUUGCUGCAAGGCUGUGCUGGAGAAGCG -.............................-..(((((((((((..((..((.(((...........))).))..))...))).)))))))).... ( -21.00, z-score = -0.51, R) >droGri2.scaffold_15203 8708551 95 - 11997470 -UCUAUCCAUCUCUCUCUCUCUCUCUCUUUAUUUGCUCAGCUGGAGUGCGGUCAUGUGUUUCAUUAUCACUGCUGCAAGGCGGUGCUGGAGAAACG -......................(((((......((.(.(((....(((((.((.(((.........)))))))))).))).).)).))))).... ( -20.10, z-score = 0.36, R) >anoGam1.chr3L 23875496 74 + 41284009 ----------------------ACGGCACCGUCGAUUCAGCUGCGCUGUGGCCACGUGUUCCACUAUCACUGCUGCAAGACGGUGCUAAUGAGACG ----------------------..(((((((((....((((.(.(..((((.........))))...).).))))...)))))))))......... ( -26.70, z-score = -1.37, R) >triCas2.ChLG3 220549 96 - 32080666 AUUGAAACGAAUCGGUUUGUGAUUGGCUGGCUGCUUUUAGCUGAAAUGCGGUCACGUGUUCCACUUACACUGCUGUAAGGCGGUGCUUAUGAAACG ......((((((((.....)))))...(((((((((((....)))).))))))))))(((.((....(((((((....)))))))....)).))). ( -23.90, z-score = -0.05, R) >consensus ______ACGAAAAUUUAUUCAUUCCUAACC_UUUUUGCAGCUGGAGUGCGGUCACGUGUUCCACUAUCAUUGCUGCAAGGCGGUGCUGGAGAAGCG ......................................((.(((((..((....))..))))))).(((..(((((...)))))..)))....... (-11.80 = -11.21 + -0.59)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:45:03 2011