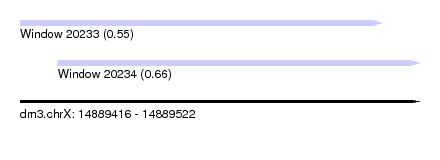
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,889,416 – 14,889,522 |
| Length | 106 |
| Max. P | 0.661411 |

| Location | 14,889,416 – 14,889,512 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 62.86 |
| Shannon entropy | 0.75025 |
| G+C content | 0.51572 |
| Mean single sequence MFE | -29.09 |
| Consensus MFE | -9.43 |
| Energy contribution | -11.76 |
| Covariance contribution | 2.33 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.554601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14889416 96 + 22422827 GAGGCUUCUAAC-UGCCACUUGGGGCAACAGUUAAG-ACAACAGAAAAUAGACGGGUUGGUGUUGGG----GGGCAGCAAUACCAACUGUUGCUCAGGUGAU ..(((.......-.)))(((((((.((((.(((...-..)))............(((((((((((..----......))))))))))))))))))))))... ( -32.30, z-score = -2.28, R) >droEre2.scaffold_4690 6629954 94 - 18748788 GAGGCUUCUAAC-UGCCACUUGGGGCAACAGUUAAG-ACAACACAAAAUAGACGGGUCUGUGGUGGG------GCAGCAGUUCCAUUUGUUGCUCAGGUGAU ..(((.......-.)))(((((((.((((((.....-..........(((((....))))).(((((------((....))))))))))))))))))))... ( -29.80, z-score = -1.06, R) >droYak2.chrX 9089921 100 + 21770863 GAGGCUUCUAAC-UGCCACUUGGGGCAACAGUUAAG-ACAACAGAAAAUAGACGGGGUGUUGGUGGGUGGAGGGCAGCAGUAUCAACUGUUGCUCAGGUGAU ...(((((((.(-..((((((.((....).(((...-..)))..........).))))...))..).))))((((((((((....)))))))))).)))... ( -28.50, z-score = -0.55, R) >droSec1.super_32 345448 96 + 562834 GAGGCUUCUAAC-UGCCACUUGGGGCAACAGUUAAA-ACAACAGAAAAUAGACGGGUUGGUGUUGGG----GCGCAGCAAUACCAACUGUUGCUCAGGUGAU ..(((.......-.)))(((((((.((((.(((...-..)))............(((((((((((..----......))))))))))))))))))))))... ( -32.30, z-score = -2.34, R) >droSim1.chrU 9432990 96 - 15797150 GAGGCUUCUAAC-UGCCACUUGGGGCAACAGUUAGG-ACAACAGAAAGUAGACGGGUUGGUGUUGGG----GGGCAGCAAUACCAACUGUUGCUCAGGUGAU ..(((.......-.)))(((((((.((((.(((...-((........)).))).(((((((((((..----......))))))))))))))))))))))... ( -33.10, z-score = -1.96, R) >droMoj3.scaffold_6473 15134456 74 - 16943266 GAGGCUUGUAACAUGCCACUUGACAUCUUGACGCUG-GUUAGAUGGGG-GGGUGGGGGGG-GAGGUGAGUGGCACGU------------------------- (..(((..(..(.(.((((((..(((((.(((....-))))))))...-)))))).)..)-...)..)))..)....------------------------- ( -20.40, z-score = -0.47, R) >droVir3.scaffold_12928 5780019 101 - 7717345 GAGGCUUGUAGCAUGCCGCUUAAGAACCUGGGGGCGUGCCACAUAACA-ACGUUAAGGAUUAAGGAUAACAAACUGUUGGCCCAACCUGUUGCACAGUCAAC ..(((((((.(((((((.((((......))))))))))).))).....-..((..(((.....((.((((.....)))).))...)))...))..))))... ( -27.20, z-score = 0.03, R) >consensus GAGGCUUCUAAC_UGCCACUUGGGGCAACAGUUAAG_ACAACAGAAAAUAGACGGGUUGGUGUUGGG____GGGCAGCAAUACCAACUGUUGCUCAGGUGAU ..(((.........)))(((((((.(((((((.....................................................))))))))))))))... ( -9.43 = -11.76 + 2.33)


| Location | 14,889,426 – 14,889,522 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 71.74 |
| Shannon entropy | 0.54068 |
| G+C content | 0.50684 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -14.11 |
| Energy contribution | -14.72 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.661411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14889426 96 + 22422827 -ACUGCCACUUGGGGCAACAGUUAAGACAACAGAAAAUAGACGGGUUGGUGUUGGG----GGGCAGCAAUACCAAC-UGUUGCUCAGGUGAUGGAACCAGGA -.((..((((((((.((((.(((.....)))............(((((((((((..----......))))))))))-)))))))))))))..))........ ( -32.30, z-score = -2.00, R) >droEre2.scaffold_4690 6629964 94 - 18748788 -ACUGCCACUUGGGGCAACAGUUAAGACAACACAAAAUAGACGGGUCUGUGGUGGG------GCAGCAGUUCCAUU-UGUUGCUCAGGUGAUGGAACCGGUA -..((((.....(((((((((...............(((((....))))).(((((------((....))))))))-)))))))).(((......))))))) ( -30.00, z-score = -0.74, R) >droYak2.chrX 9089931 100 + 21770863 -ACUGCCACUUGGGGCAACAGUUAAGACAACAGAAAAUAGACGGGGUGUUGGUGGGUGGAGGGCAGCAGUAUCAAC-UGUUGCUCAGGUGAUGGAACCGGUA -.(..((.((((.(....)...)))).(((((..............)))))...))..).((((((((((....))-)))))))).(((......))).... ( -30.24, z-score = -0.95, R) >droSec1.super_32 345458 96 + 562834 -ACUGCCACUUGGGGCAACAGUUAAAACAACAGAAAAUAGACGGGUUGGUGUUGGG----GCGCAGCAAUACCAAC-UGUUGCUCAGGUGAUGGAAACAGGA -.....((((((((.((((.(((.....)))............(((((((((((..----......))))))))))-))))))))))))).((....))... ( -32.80, z-score = -2.80, R) >droSim1.chrU 9433000 96 - 15797150 -ACUGCCACUUGGGGCAACAGUUAGGACAACAGAAAGUAGACGGGUUGGUGUUGGG----GGGCAGCAAUACCAAC-UGUUGCUCAGGUGAUGGAAACAGGA -.....((((((((.((((.(((...((........)).))).(((((((((((..----......))))))))))-))))))))))))).((....))... ( -33.60, z-score = -2.56, R) >droVir3.scaffold_12928 5780029 96 - 7717345 GCAUGCCGCUUAAGA-ACCUGGGGGCGUGCCACAUAACAACGUUAAGGAUUAAGGAUAACAAACUGUUGGCCCAACCUGUUGCACAGUCAACAUCAG----- (((((((.((((...-...)))))))))))...........((..(((.....((.((((.....)))).))...)))...))..............----- ( -21.20, z-score = 0.82, R) >consensus _ACUGCCACUUGGGGCAACAGUUAAGACAACAGAAAAUAGACGGGUUGGUGUUGGG____GGGCAGCAAUACCAAC_UGUUGCUCAGGUGAUGGAACCAGGA ..((..((((((((.((((((((.....)))...................(((((................))))).)))))))))))))..))........ (-14.11 = -14.72 + 0.62)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:45:01 2011