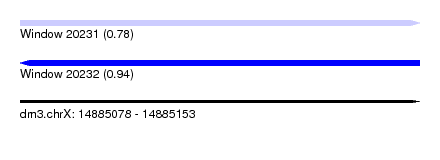
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,885,078 – 14,885,153 |
| Length | 75 |
| Max. P | 0.935087 |

| Location | 14,885,078 – 14,885,153 |
|---|---|
| Length | 75 |
| Sequences | 6 |
| Columns | 80 |
| Reading direction | forward |
| Mean pairwise identity | 73.73 |
| Shannon entropy | 0.50688 |
| G+C content | 0.56388 |
| Mean single sequence MFE | -20.20 |
| Consensus MFE | -15.71 |
| Energy contribution | -15.27 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.65 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.775410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
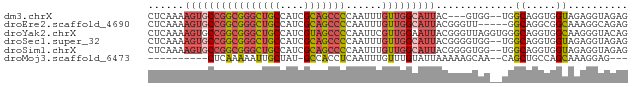
>dm3.chrX 14885078 75 + 22422827 CUCUACCUCUACCACCUGCCA--CCAC---GUAAUGCCAACAAAUUGGGGCUGCGAUGGCAGCCCGCCGGCACUUUUGAG ......(((.......((((.--....---.((((........))))(((((((....)))))))...)))).....))) ( -21.90, z-score = -1.35, R) >droEre2.scaffold_4690 6625695 75 - 18748788 CUCUGCCUUUGCCGCCUGCC-----AACCCGUAAUGCCAACAAAUUGGGGCUGCGAUGGCAGCCCGCCGGCACUUUUGAG (((......(((((((((((-----(...((((.(.((((....)))).).)))).))))))...).))))).....))) ( -24.60, z-score = -0.51, R) >droYak2.chrX 9084824 80 + 21770863 CUGUACCCUUGCCACCUGCCCACCUAACCCGUAAUUCCAACGAAUUGGGGCUACGAUGGCAGCCCGCCGGCACUUUUGAG .........((((..(((((....((.(((.((((((....))))))))).))....)))))......))))........ ( -19.30, z-score = -0.28, R) >droSec1.super_32 341245 78 + 562834 CUCUACCUCUACCACCUGCCA--CCACCCCGUAAUGCCAACAAAUUGGGGCUGCGAUGGCAGCCCGCCGGCACUUUUGAG ......(((.......((((.--........((((........))))(((((((....)))))))...)))).....))) ( -21.90, z-score = -1.23, R) >droSim1.chrX 11487588 78 + 17042790 CUCUACCUCUACCACCUGCCA--CCACCCCGUAAUGCCAACAAAUUGGGGCUGCGAUGGCAGCCCGCCGGCACUUUUGAG ......(((.......((((.--........((((........))))(((((((....)))))))...)))).....))) ( -21.90, z-score = -1.23, R) >droMoj3.scaffold_6473 15127611 64 - 16943266 ---CUCCUUUGCUGGCAGCUG--UUGCUUUUUAAUACAAACAAAUUGAGGUGGC-AUAGCAAUUUUUGAG---------- ---(((..((((((.....((--(..((((................))))..))-))))))).....)))---------- ( -11.59, z-score = 0.70, R) >consensus CUCUACCUCUACCACCUGCCA__CCACCCCGUAAUGCCAACAAAUUGGGGCUGCGAUGGCAGCCCGCCGGCACUUUUGAG ................((((...........((((........))))(((((((....)))))))...))))........ (-15.71 = -15.27 + -0.44)
| Location | 14,885,078 – 14,885,153 |
|---|---|
| Length | 75 |
| Sequences | 6 |
| Columns | 80 |
| Reading direction | reverse |
| Mean pairwise identity | 73.73 |
| Shannon entropy | 0.50688 |
| G+C content | 0.56388 |
| Mean single sequence MFE | -27.92 |
| Consensus MFE | -16.95 |
| Energy contribution | -17.70 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.935087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
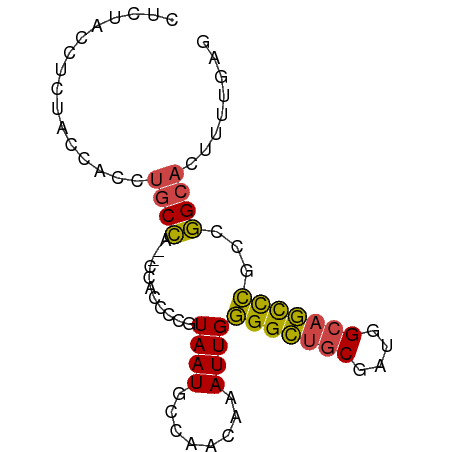
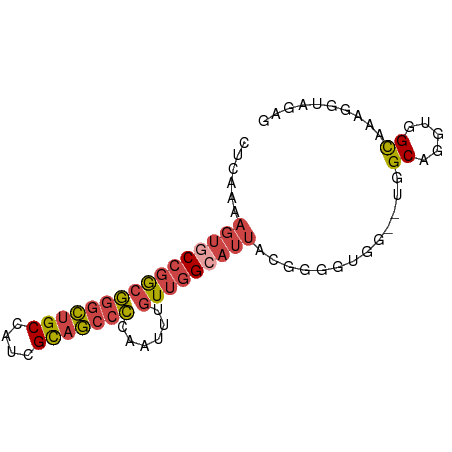
>dm3.chrX 14885078 75 - 22422827 CUCAAAAGUGCCGGCGGGCUGCCAUCGCAGCCCCAAUUUGUUGGCAUUAC---GUGG--UGGCAGGUGGUAGAGGUAGAG (((......(((((((((((((....)))))))......))))))(((((---.((.--...)).))))).)))...... ( -28.90, z-score = -1.53, R) >droEre2.scaffold_4690 6625695 75 + 18748788 CUCAAAAGUGCCGGCGGGCUGCCAUCGCAGCCCCAAUUUGUUGGCAUUACGGGUU-----GGCAGGCGGCAAAGGCAGAG (((.....(((((.((((((((....))))))).....(((..((.......)).-----.)))).)))))......))) ( -32.40, z-score = -1.75, R) >droYak2.chrX 9084824 80 - 21770863 CUCAAAAGUGCCGGCGGGCUGCCAUCGUAGCCCCAAUUCGUUGGAAUUACGGGUUAGGUGGGCAGGUGGCAAGGGUACAG .......(((((.((.(.(((((..(.((((((.(((((....)))))..)))))).)..))))).).))...))))).. ( -35.00, z-score = -3.23, R) >droSec1.super_32 341245 78 - 562834 CUCAAAAGUGCCGGCGGGCUGCCAUCGCAGCCCCAAUUUGUUGGCAUUACGGGGUGG--UGGCAGGUGGUAGAGGUAGAG ........((((.((.(.((((((((...(((((....((....))....)))))))--)))))).).))...))))... ( -31.60, z-score = -1.78, R) >droSim1.chrX 11487588 78 - 17042790 CUCAAAAGUGCCGGCGGGCUGCCAUCGCAGCCCCAAUUUGUUGGCAUUACGGGGUGG--UGGCAGGUGGUAGAGGUAGAG ........((((.((.(.((((((((...(((((....((....))....)))))))--)))))).).))...))))... ( -31.60, z-score = -1.78, R) >droMoj3.scaffold_6473 15127611 64 + 16943266 ----------CUCAAAAAUUGCUAU-GCCACCUCAAUUUGUUUGUAUUAAAAAGCAA--CAGCUGCCAGCAAAGGAG--- ----------(((.....(((((..-((...((....((((((........))))))--.))..)).)))))..)))--- ( -8.00, z-score = 1.04, R) >consensus CUCAAAAGUGCCGGCGGGCUGCCAUCGCAGCCCCAAUUUGUUGGCAUUACGGGGUGG__UGGCAGGUGGCAAAGGUAGAG ......((((((((((((((((....)))))))......))))))))).............((.....)).......... (-16.95 = -17.70 + 0.75)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:45:00 2011