
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,852,759 – 14,852,886 |
| Length | 127 |
| Max. P | 0.757031 |

| Location | 14,852,759 – 14,852,886 |
|---|---|
| Length | 127 |
| Sequences | 5 |
| Columns | 155 |
| Reading direction | forward |
| Mean pairwise identity | 51.42 |
| Shannon entropy | 0.84536 |
| G+C content | 0.42024 |
| Mean single sequence MFE | -38.86 |
| Consensus MFE | -11.97 |
| Energy contribution | -11.09 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.96 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.757031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
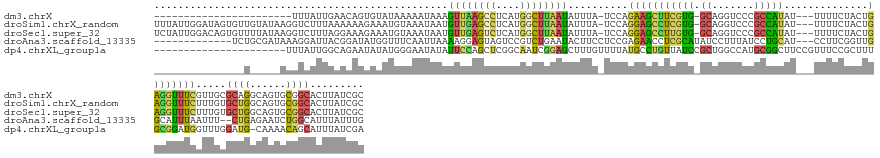
>dm3.chrX 14852759 127 + 22422827 -----------------------UUUAUUGAACAGUGUAUAAAAAUAAAGUUAAGCCUCAUGGCUUAAUAUUUA-UCCAGAAGCUUCGUG-GCAGGUCCCGCCAUAU---UUUUCUACUGAGGUUUCGUUGCGCAGGCAGUGCGGCACUUAUCGC -----------------------...........(((.((((..(((((((((((((....)))))))).))))-)...(((((((((((-((.(....))))))..---.........))))))))(((((((.....)))))))..))))))) ( -35.60, z-score = -1.67, R) >droSim1.chrX_random 3894109 150 + 5698898 UUUAUUGGAUAGUGUUGUAUAAGGUCUUUAAAAAAGAAAUGUAAAUAAUGUUGAGCCUCAUGGCUUAAUAUUUA-UCCAGGAGCCUCGUG-GCAGGUCCCGCCAUAU---UUUUCUACUGAGGUUUCUUUGUGCUGGCAGUGCGGCACUUAUCGC .......(((((((((((((...(((......((((((((.(....(((((((((((....)))))))))))..-..(((..(((....)-)).((.....))....---.......)))).)))))))).....))).))))))))).)))).. ( -46.20, z-score = -2.66, R) >droSec1.super_32 308044 150 + 562834 UCUAUUGGACAGUGUUUUAUAAGGUCUUUAGGAAAGAAAUGUAAAUAAUGUUGAGUCUCAUGGCUUAAUAUUUA-UCCAGGAGCCUUGUG-GCAGGUCCCGCCAUAU---UUUUCUACUGAGGUUUCUUUGUGCUGGCAGUGCGGCACUUAUCGC ((((..((((..(((...)))..)))).))))((((((((.(....(((((((((((....)))))))))))..-..(((.((...((((-((.(....))))))).---....)).)))).))))))))((((((......))))))....... ( -40.80, z-score = -0.79, R) >droAna3.scaffold_13335 2510416 137 + 3335858 -------------UCUGCGAUAAAGGAUUACGGAUAUGGUUUCAAUUAAAAGGAGUAGUCCGUCUGAAUACUUCCUCCGAGAACCUCGCAUAUCCUUUAUCCUGCAU---CCUUCGGUUGGCAUUUAAUUU--CUGAGAAUCUGGCAUUUAUUUG -------------..((((((..(((((...(((((((............(((((((.((.....)).))).)))).((((...)))))))))))...)))))..))---).(((((..(........)..--)))))......)))........ ( -30.00, z-score = -0.01, R) >dp4.chrXL_group1a 5889504 132 + 9151740 ----------------------UUUAUUGGCAGAAUAUAUGGGAAUAUAUUCCAGCUCGGCAAUCGGAGCUUUGUUUUAUGCCUGUUAUCCGCUGGCCAUGCGGCUUCCGUUUCCGCUUUGCGGAUGGUUUGGAUG-CAAAACAGCAUUUAUCGA ----------------------......((((((((((((....)))))))).(((((.(....).)))))........))))..((((((((.(((...((((...))))....)))..))))))))..((((((-(......))))))).... ( -41.70, z-score = -1.12, R) >consensus _______________U_____AGGUCUUUAAAAAAGAAAUGUAAAUAAAGUUGAGCCUCAUGGCUUAAUAUUUA_UCCAGGAGCCUCGUG_GCAGGUCCCGCCAUAU___UUUUCUACUGAGGUUUCGUUGUGCUGGCAGUGCGGCACUUAUCGC .................................................((((((((....))))))))..........(((((((((((.((.......)))))..............)))))))).....((((......))))......... (-11.97 = -11.09 + -0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:44:53 2011